1. Introduction
Beta-lactam has been an efficient antibiotic that has been frequently used for the treatment of infections for many years. However, bacterial resistance to this antibiotic has increased gradually due to the transfer of beta-lactamase-encoding genes, especially through conjugation. This has recently caused the resistance of bacteria to beta-lactam antibiotics (1, 2). Extended-spectrum beta-lactamases (ESBLs) are a group of beta-lactamase enzymes that are able to hydrolyze oximinocephalosporins (3). Two of these enzymes are TEM and SHV. They are productions of point mutations in the main inactive extended-spectrum enzymes. In recent years, nonderivative beta-lactamases of TEM and SHV have been reported, and most of them have been CTX-M enzymes (4-6).
In 1989, the CTX-M family of ESBLs was reported in Germany for the first time, and later, these ESBLs spread to different parts of the world. The CTX-M enzyme has been found frequently in Escherichia coli and Klebsiella, but it has also been reported in other Enterobacteriaceae species (3). Generally, plasmids are responsible for the gene development of beta-lactamases among similar or different Enterobacteriaceae strains. Some strains of Enterobacteriaceae produce beta-lactamases that are encoded by chromosomes and that could cause antibiotic resistance (7, 8).
Studies have indicated that more than 60 types of CTX-M enzymes have been identified. These enzymes are categorized into five main groups based on amino acid variations (9). In comparison to TEM and SHV beta-lactamases, CTX-M has a more harmful effect on cefotaxime and ceftriaxone than ceftazidime. The CTX-M enzyme is inhibited by beta-lactamases, such as clavulanic acid, sulbactam, and tazobactam (10, 11).
2. Objectives
Since blaCTX-M and blaCTX-M-1 are common genes among ESBL producers (12), this study aimed to investigate the frequency of the beta-lactamase CTX-M gene in Enterobacteriaceae strains that have been isolated from hospitalized patients in the teaching hospitals of Ahvaz, Iran. The results of this study could help physicians choose more effective antibiotics for treating patients.
3. Methods
3.1. Bacteria Isolation from Specimens
In this study, Enterobacteriaceae isolates were collected from the Imam Khomeini and Golestan teaching hospitals in Ahvaz, Iran over the course of 11 months from April 2012 to February 2013. These bacteria were isolated from wounds (n = 5), blood (n = 10), urine (n = 210), trachea (n = 10), and fluids (n = 2) and were transported to the microbiology lab at the school of medicine. The isolates were identified using standard biochemical tests, such as triple sugar iron agar, lysine urea, SIM, MR-VP, citrate, and gas production testing (all the culture mediums were provided by Merck, Germany) (13).
3.2. Phenotypic ESBLs with The Combination Disk Method
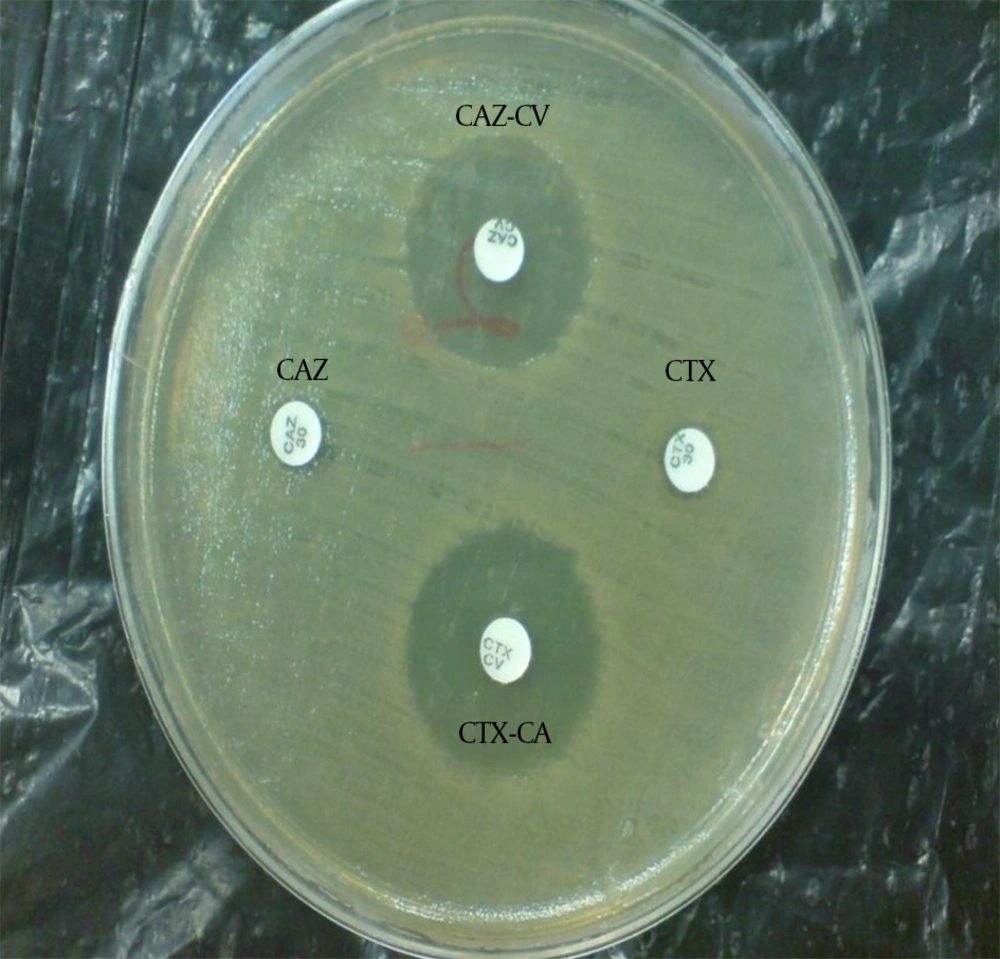
The ESBL-producing Enterobacteriaceae were identified using the phenotypic combination disk method based on CLSI directions (13). With this method, combination disks of cefotaxime-clavulanic acid (CTX30-CA10) and ceftazidime-clavulanic acid (CAZ30-CA10) with single disks of cefotaxime and ceftazidime (Mast company, England) in Mueller-Hinton agar (Merck, Germany) were used. First, a microbe suspension equal with the McFarland 0.5 standard was prepared and was thoroughly spread in the culture medium. Then, paper disks of the antibiotics were placed on the plate in 2.5 cm apart from each other. The plates were incubated at 35°C for 18 - 24 hours. Then, the bacterial sensitivity or resistance to the antibiotics were examined. If the inhibition zone around the CAZ-CA and CTX-CA combination disks were at least 5 mm more than the single disks, a tested isolate was considered to be an ESBL-producing strain (Figure 1).
CAZ, ceftazidime; CAZ-CV, ceftazidime-clavulanic acid; CTX, cefotaxime; CTX-CV, cefotaxime-clavulanic acid; No growth inhibition zone around the single disk of ceftazidime and cefotaxime indicates bacterial resistance. The growth inhibition zone around the ceftazidime and cefotaxime disks in combination with clavulanic acid shows the sensitivity of the bacteria to the combination disks and confirms the production of ESBLs by the isolate.
3.3. DNA Extraction and PCR of the CTX-M and CTX-M-1 Genes
DNA extraction was performed using the boiling method (14). In this method, three to four colonies of positive phenotypic strains were suspended in microtubes with 500 µL of sterile distilled water. They were placed in a thermo black machine for ten minutes at 100°C. Then, the samples were centrifuged for 10 minutes at 4°C and 12000 rpm. In addition, 200 µL of the surface fluid was separated in a sterile condition and was stored in Eppendorf micro tubes at -20°C. The concentration of the extracted DNA was measured using a BioPhotometer (Eppendorf, Germany).
The PCR reactions were carried out using two pairs of primers: CTX-M and CTX-M-1 genes (microbiology biotechnology gene). The sequences of these primers are presented in Table 1. For each PCR reaction, a master mix of 25 µL that included 2.5 µL of PCR buffer 10X, 0.5 µL of 10 mM dNTP mix, 0.75 µL of 50 mM MgCl2, 0.2 µL of 5u/µL Taq DNA polymerase, 2 µL of template DNA, 2 µL of 10 µM F and R primers, and 17.05 µL of distilled water was prepared (15, 16).
The best conditions for the PCR reaction were recognized in a pilot method by performing multiple repetitions of the test and concentration changes and the temperature gradient in a Master Cycler (Ependorff, Germany). The program of gene amplification in the Master Cycler consisted of an initial denaturation at 94°C for 5 minutes, annealing at 55°C for 45 seconds, 30 cycles, an extension at 72°C for 1 minute, 30 cycles, and a final extension for and 5 minute, one cycle.
3.4. PCR Production Electrophoresis
PCR production was evaluated using 1.2% gel agarose with the addition of 0.5 µg/mL ethidium bromide (CinaGen Co., Iran). The photography of the related gel was carried out using Gel documentation (Protein Simple Co., US). The standard strains of E. coli ATCC 25922 and an E. coli strain possessing the CTX-M gene, which was determined by sequencing, were used as negative and positive controls, respectively.
4. Results
The isolation results of the different types of Enterobacteriaceae isolates are summarized in Table 2. Based on these results, the most frequent isolates were E. coli (71.3%), Enterobacter (27%), and Klebsiella (1.2%). In addition, the phenotypic investigation of the ESBLs in the 240 Enterobacteriaceae isolates indicated that 108 isolates (45%) were producers of ESBLs and that most of these producers (79 isolates) were E. coli (Table 3). However, the PCR results confirmed that 104 strains of the ESBL-producing isolates (96%) had the CTX-M gene and 4 isolates did not have this gene.
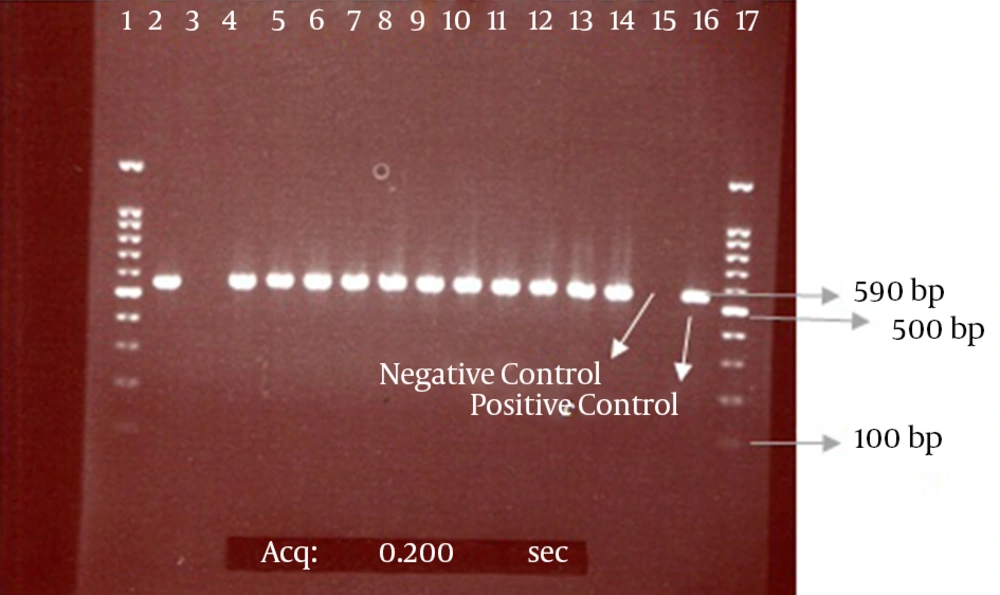
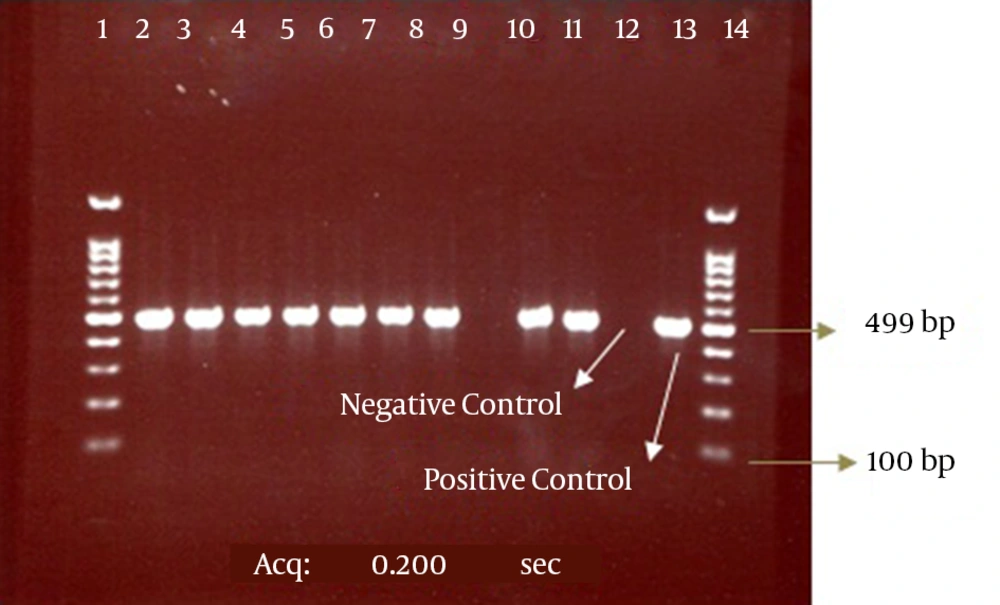
Our results also showed that among 108 positive phenotype ESBL isolates, 99 isolates (92%) had the CTX-M-1 gene and 9 isolates lacked this gene according to the results of the PCR method. The electrophoresis of the PCR products for the CTX-M and CTX-M-1 genes are presented in the Figures 2 and 3.
| Bacterial Names | No. (%) of Isolates |
|---|---|
| E. coli | 171 (71.3) |
| K. pneumoniae | 2 (0.8) |
| K. oxytoca | 1 (0.4) |
| E. aerogenes | 19 (8) |
| E. cloacae | 5 (2) |
| E. taylore | 2 (0.8) |
| E. intermedium | 1 (0.4) |
| E. gergoviae | 38 (15.9) |
| C. freundii | 1 (0.4) |
| Total | 240 (100) |
| Name | ESBL+, No. (%) | ESBL-, No. (%) | Total Isolates, No. (%) |
|---|---|---|---|
| E. coli | 79 (46.1) | 92 (53.9) | 171 (100) |
| K. pneumoniae | 0 (0) | 1 (100) | 1 (100) |
| K. oxytoca | 2 (100) | 0 (0) | 2 (100) |
| E. aerogenes | 8 (42.1) | 11 (57.9) | 19 (100) |
| E. cloacae | 5 (100) | 0 (0) | 5 (100) |
| E. taylore | 0 (0) | 2 (100) | 2 (100) |
| E. intermedium | 0 (0) | 1 (100) | 1 (100) |
| E. gergoviae | 14 (36.9) | 24 (63.1) | 38 (100) |
| C. freundii | 0 (0) | 1 (100) | 1 (100) |
| Total | 108 (45) | 132 (55) | 240 (100) |
The bond 590 bp is related to the CTX-M beta-lactamase gene. Lanes 1 and 17 contain a 100-bp DNA size marker; Lane 3, CTX-M-negative isolate; Lanes 2 and 4 - 14, CTX-M-positive isolates; Lanes 15 and 16, negative (E. coli ATCC 25922) and positive (E. coli strain possessing the CTX-M gene, which was determined by sequencing) controls, respectively.
The bond 499 bp is related to the CTX-M-1 beta-lactamase gene. Lanes 1 and 14, DNA size marker; Line 9, CTX-M-1-negative isolate; Lanes 2 - 8, 10, and 11, CTX-M-1-positive isolates; Lanes 12 and 13, negative (E. coli ATCC 25922) and positive (E. coli strains possessing the CTX-M-1 gene, which was determined by sequencing) controls, respectively.
5. Discussion
Beta-lactam antibiotics are the first medicine option for the treatment of infections due to the Gram-negative bacilli of Enterobacteriaceae. However, in the past decade, bacterial resistance to these antibiotics has increased (17, 18). The main mechanism of resistance among Enterobacteriaceae members to beta-lactam antibiotics is the production of beta-lactamases enzymes (18). ESBL enzymes were separated from Klebsiella pneumoniae for the first time in Germany in 1983, but today, nasocomial infections due to infections by ESBL-producing organisms have developed all over the world (19, 20).
Among the ESBLs produced by Enterobacteriaceae isolates, the CTX-M family has had the most destructive impact on cefotaxime and ceftazidime. The appropriate phenotypic method for the detection of beta-lactamase-producing bacteria is the combination disk method according to CLSI directions. With this method, beta-lactamase activity is inhibited by clavulanic acid, so the growth inhibition zone around the combination disk is increased to a single antibiotic disk, which is a criteria for the detection of ESBL isolates (13). The results of our study on Enterobacteriaceae isolates showed that 108 isolates (45%) were phenotypic ESBL producers, and the most frequent producers belonged to the Klebsiella (66.6%), E. coli (46.1%), and Enterobacter (41.5%) isolates.
Some studies in different countries reported various frequencies of ESBLs among Enterobacteriaceae isolates. For instance, a study carried out on Enterobacteriaceae isolates by Luzzaro et al. (2006) in Italy showed that 7.4% of isolates were phenotypic ESBL producers and that the most frequent ESBL producers were E. coli strains (31.9%) (21). In addition, in a study on Enterobacteriaceae isolates in Pakistan, Riaz et al. (2011) showed that among 1018 isolates, 300 (29.5%) isolates were phenotypic ESBL producers and that the frequency of E. coli and Klebsiella among these ESBL producers was (39.3%) and (26.1%), respectively (22).
Studies have also been conducted on this subject in Iran. In a study in Sannandaj, Ramezanzadeh et al. (2010) revealed that 14.5% of Gram-negative bacilli were ESBL-producing isolates (23). In addition, in a study carried out by Moosavian et al. (2012) in Dezful, Iran, the ESBL rate in Enterobacteriaceae isolates was 30.5%, and the prevalence of K. pneumoniae and E. coli among the ESBL producers in this study was 45.4% and 28.8%, respectively (24). Moreover, in a study on Klebsiella isolates in Tehran, Nasehi et al. (2010) showed that the ESBL frequency was 38.5 %, which was lower than the present study’s result (25).
The results discrepancy between the present study and these other studies might be due to the consumption patterns of antibiotics, particularly cephalosporins; the number of antibiotic prescriptions; and the differences in the time periods during which the Enterobacteriaceae isolates were collected (4). The results of our study showed that among 108 phenotypically positive Enterobacteriaceae isolates, 104 isolates (96%) had the CTX-M gene and 4 isolates did not have this gene. The frequency of the CTX-M gene in Klebsiella was 66.6%, but in E. coli and Enterobacter, this frequency was 46.1% and 41.5%, respectively. In addition, among 108 phenotypically positive Enterobacteriaceae isolates, 99 (92%) isolates had the CTX-M-1 gene and 5 isolates lacked the gene. The nonexistence of the CTX-M and CTX-M-1 genes in strains that were phenotypically positive in the combination disk test revealed that these strains can have other ESBL enzymes, such as TEM and SHV (26).
Maina et al. (2012) reported that among 52 phenotypically positive Enterobacteriaceae isolates in Kenya, 46 (88.5%) had the CTX-M gene but 6 isolates lacked the gene (26). The reported frequencies of this gene in was 96% for E. coli and 79% for K. pneumoniae, which are consistent with the results of the present study in Ahvaz. Furthermore, the frequency of the CTX-M gene in Peirano et al.’s study on E. coli isolates in Turkey was reported to be 63.9%. (27). In Iran, there also have been studies that indicated the frequency of ESBL-producing Enterobacteriaceae isolates, such as CTX-M.
Soltan Dalal et al. (2011) studied 188 isolates of E. coli strains collected from Tabriz hospitals in Iran which indicated 84.1% of strains were positive for CTX-M-1 (28). The results of studies by Shahcheraghi et al. (2010) and Mirzaiee et al. (2009) concerning isolates from Tehran reported the frequencies of the CTX-M gene in E. coli isolates to be 20.8% and 35.7%, respectively (16, 29). Although cephalosporins are effective for treating Enterobacteriaceae infections, different studies have indicated the regionally high frequency of ESBLs, especially CTX-M. Since CTX-M genes are associated with the appended sequential of ISEcp1 and since these sequences are responsible for the movement and expression of the CTX-M gene, the transfer and movement of appended ISEcp1 sequences between different strains motivates the expression of the CTX-M gene and the subsequent epidemic development of ESBLs (30). The transferability of these genes from animal sources to humans is considered to be one of the important causes of the high prevalence of the CTX-M gene (31).
5.1. Conclusion
The results of this study phenotypically and genotypically confirmed the high frequency of ESBL-producing strains among Enterobacteriaceae isolates, such as the xCTX-M and CTX-M-1 genes, in our region. Therefore, conducting antibiotic susceptibility tests for the detection of ESBL isolates prior to prescribe of beta-lactam antibiotics is recommended. Conducting these tests could help prevent the spread of strains that are resistant to beta-lactam antibiotics.