1. Background
Chronic wounds affect 1 - 2% of the population in the entire world, causing persistent morbidity with frequent delay in the healing process and a high recurrence rate (1). Delay of wound healing is becoming one of the major problems imposing heavy burden on public health, and its common factor is the presence of bacterial flora on chronic wounds (2). Pseudomonas aeruginosa is the most common microorganism isolated from chronic wounds, often found as biofilm producer, that acts as a barrier in wound healing and shows resistance to antimicrobial therapy (2-4). Pseudomonas aeruginosa is a Gram-negative and rod-shaped opportunistic pathogen that is a major causative microorganism in wound infections, delaying the wound healing process.
Chronic wounds are classified in different categories, such as diabetic foot ulcer (DFU), venous leg ulcers (VLU), pressure ulcer (PU), surgical site infection (SSI), and abscess or trauma ulcers (5). These types of infections are prolonged and become chronic because of bacterial contribution to chronic wounds, on which biofilms develop and increase the normal period of healing (6, 7). Biofilm is actually a structured consortium of bacteria and extracellular matrix, which is essential for interconnecting the bacteria and can be composed of polysaccharides, proteins, and extracellular DNA (eDNA) (8). It protects bacterial cells from host defense syatem (9) and impairs healing (10). The existence of biofilms in wounds has been reported in vivo animal data and in vitro models. Sixty percent of chronic wound specimens were characterized as biofilm-containing, whereas only 6% of acute wounds contained biofilm, indicating biofilms were prevalent in chronic wound samples while relatively rare in samples from acute wounds (11, 12).
Microbial involvement in the production of biofilms in chronic wounds was reviewed recently (13). Pseudomonas aeruginosa was the fourth most frequently isolated pathogen in chronic wound infections (8%), and the seventh leading contributor to bloodstream infections (2 to 6%) (14). Epidemiological outcome studies have shown that infections caused by drug-resistant P. aeruginosa could be associated with significant increases in morbidity, mortality, the need for surgical intervention, the length of hospital stay and chronic care, and overall cost of treating the infection (15). Chronic wounds are mostly found in diabetic patients, and the mortality rate is 39 - 80% in patients after the development of diabetic foot ulcer (16). Pseudomonas aeruginosa exhibits the highest rates of resistance to fluoroquinolones, with resistance to ciprofloxacin and levofloxacin ranging from 20 to 35%. Gentamicin was the least active of the aminoglycosides, with lower rates of resistance being reported for tobramycin and amikacin in most studies (17, 18). The present study was designed to discover the prevalence, isolation, and identification of P. aeruginosa and its biofilm production in chronic wounds and its resistance to antibiotics in different types of chronic wounds. This study was significant to overcome the problem of the drug of choice in chronic wound patients and to prevent the patients from over use of drugs.
2. Objectives
The present study was designed to evaluate the prevalence of P. aeruginosa and its antibiotics resistance in different types of chronic wounds. This study was significant to overcome the problem of the drug of choice in chronic wound patients.
3. Methods
3.1. Ethics Statement
The research was approved by the departmental and University ethics committee No. Kust173638. All data given from patients were full with the agreement of patients and hospital committee.
3.2. Sampling
This study was conducted from September 2014 to June 2015. A total of 91 pus or wound swabs from different patients with chronic wound infections including diabetic foot ulcers, non-healing surgical wounds, venous leg ulcers, leg ulcers, and non-healing wounds from an abscess or trauma were collected from different wards (OPD, Medical, Orthopaedic, Burns, Surgical Ward, and Main Operating Theatre) from DHQ Hospital KDA District, Kohat (Khyber Pakhtunkhwa), Pakistan. The isolates of chronic wounds were transported by Cary-Blair Medium (Oxoid Ltd. UK). These wound exudates were immediately transported to the laboratory of the Department of Microbiology, Kohat University of Science and Technology, Pakistan, for further processing.
3.3. Isolation of P. aeruginosa from Clinical Samples
Pseudomonas aeruginosa was isolated from each sample as per the provided protocol. Each sample was inoculated onto pseudomonas isolation agar (PIA) followed by identification of P. aeruginosa based on Gram staining reaction and biochemical tests such as Oxidase, Motility, Citrate, and Catalase, while negative for Methyl Red, Voges-Proskauer, Urease and Indole tests, as discussed previously by Dortet et al. (19).
3.4. Antimicrobial Susceptibility Testing
The antibiotic resistance patterns of P. aeruginosa were assessed using the Kirby-Bauer disk diffusion method, following the recommendations of the clinical and laboratory standard institute (CLSI) (20). Muller-Hinton agar was used to check the resistance of isolates to antibiotic discs. A 0.5 McFarland turbidity standard equivalent bacterial suspension for inoculation was prepared and inoculated. Antibiotic disks (Oxoid Ltd. UK) were applied, and the plate was incubated at 37°C for 48 hours. Amikacin (30 μg), Augmentin (30 μg), Cefepime (30 µg), Ceftriaxone (30 μg), and Ciprofloxacin (5 μg) were used in the present study.
3.5. Biofilm Assay
Biofilm formation was determined by plate assay, as previously reported by Freeman et al. (21). Briefly, bacterial isolates were cultured on Congo red agar (CRA agar) plates and then incubated for 24 - 48 hours at 37°C.
3.6. PCR (Polymerase Chain Reaction)
3.6.1. Detection of the algD GDP-Mannose Dehydrogenase Gene
The algD GDP-mannose dehydrogenase gene of P. aeruginosa contains 2032 bp (GenBank, AC. No. 400337, Identification No. g45267). The selected primers VIC 1 (5'TTCCCTCGCAGAGAAAACATC3') and VIC2 (5'CCTGGTTGATCAGGTCGATCT 3') were designed to amplify a 520-bp segment of the algD GDP-mannose dehydrogenase gene of P. aeruginosa. To assure DNA quality, universal bacterial primers 1 lE-l3B were used to target the 16 S RNA gene of the bacterium (22).
3.7. Statistical Analysis
The results were analyzed using SPSS version 16 software. P-values <0.05 were considered significant.
4. Results
A total of 91 samples were collected from patients with different ulcers, including diabetic ulcers, surgical ulcers, venous leg ulcers, pressure ulcers, and abscess or trauma ulcers (as shown in Table 1). Both genders were included in the study, and more ulcers were observed in males (N = 63; 69%) than in females (N = 28; 31%) with a statistically non-significant association (P > 0.05). The frequency of diabetic ulcers was found to be highest (43.96%) followed by the frequency of surgical ulcers (19.78%) (Table 1).
| Types of Wound | Total (n = 91) | Male (n = 63) | Female (n = 28) | P Value |
|---|---|---|---|---|
| Diabetic ulcers | 40 (43.96) | 31 (49.21)a | 09 (32.14) | 0.657 (P > 0.050) |
| Surgical ulcers | 18 (19.78) | 11 (17.47) | 07 (25) | |
| Venous leg ulcers | 14 (15.38) | 09 (14.29) | 05 (17.86) | |
| Pressure ulcers | 10 (10.99) | 06 (9.52) | 04 (14.29) | |
| Abscess or trauma ulcers | 9 (9.89) | 06 (9.52) | 03 (10.71)a | |
| Total | 91 | 63(69) | 28 (31) |
Gender-Wise Lesions Percentage and P Value of Different Samples
4.1. Phenotypic Results
Of the 91 samples collected from different types of wounds, 44 (48.3%) isolates were identified as P. aeruginosa, which were further confirmed through a series of different biochemical tests. The results showed this bacterium was positive for Oxidase, Motility, Citrate, and Catalase, while negative for Methyl Red, Voges-Proskauer, Urease, and Indole Tests (Table 2). The highest percentage of P. aeruginosa (62.5%) was found in diabetic ulcers, followed by venous leg ulcers (50%), with a statistically non-significant association (P > 0.05) (Table 3).
| Types of Wounds | P. aeruginosa | P Value | ||
|---|---|---|---|---|
| Positive | Negative | Percentage | ||
| Diabetic ulcers | 25 | 15 | 62.5 | 0.1042 (P > 0.05) |
| Surgical ulcers | 07 | 11 | 38.8 | |
| Venous leg ulcers | 07 | 07 | 50 | |
| Pressure ulcers | 03 | 07 | 27.2 | |
| Abscess or trauma ulcers | 02 | 07 | 22.2 | |
| Total (%) | 44 | 47 | 48.3 | |
Isolation of P. aeruginosa from Different Ulcers
4.2. Molecular Identification and Biofilm Production
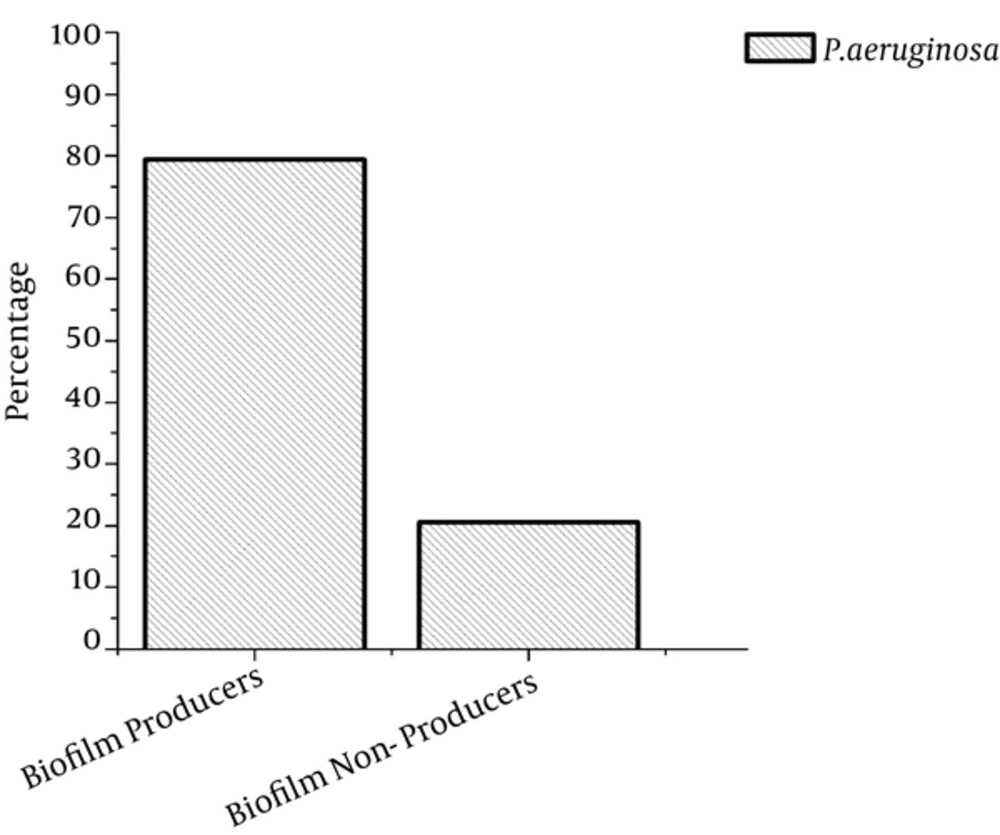
PCR was used for further confirmation, targeting the algD gene (520 bp) because it is one of the conserved genes in P. aeruginosa (Figure 1). After PCR, the confirmed P. aeruginosa isolates were further subjected to an evaluation of biofilm production, and 79.5% of samples were found to produce biofilm, while 20.5% did not produce biofilm (Figure 2).
4.3. Antibiotic Sensitivity Patterns
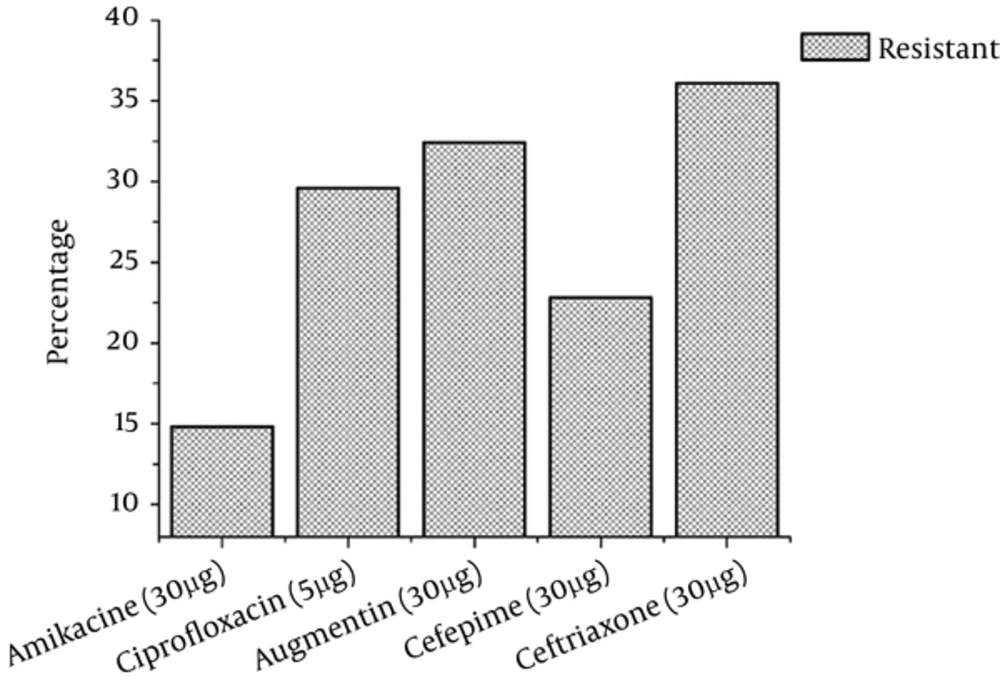
The antibiotic sensitivity patterns of P. aeruginosa isolated from clinical specimens were found to be variable. P. aeruginosa isolates were more resistant to ceftriaxone (30 µg) followed by Augmentin (30 µg), ciprofloxacin (5 μg), Cefepime (30 µg), and amikacin (30 µg). The amikacin had a very positive action indicating that P. aeruginosa was very less resistant to it. The resistance frequency of the pathogen to different antibiotics is shown in Figure 3.
5. Discussion
Rupture of skin due to any accidental cut, bite, or burn is called wound. Every wound has its own period to heal. When the wound does not heal at its specific period, it becomes chronic. The chronicity of wounds depends on many factors, the major of which is bacterial contribution as reviewed by Rahim et al. in 2016 (2). Chronic wounds contain diverse microbial flora that form biofilm in the wound, further increasing bacterial resistance to drugs. The current findings showed P. aeruginosa is present in most of the chronic wounds and the highest prevalence was found in diabetic patients. Similarly, some studies reported P. aeruginosa is the most common pathogen in chronic wounds, and found that it is very problematic due to its ability to form biofilms that are highly resistant to antimicrobial agents (23-27).
Recently in a study, diabetic mouse model was used to determine the ability of the opportunistic pathogen, P. aeruginosa, to cause biofilm-associated infections, showing the highest risk of chronic infection in the diabetic model (28). It is an opportunistic pathogen, frequently acquired in hospital environments and often is associated with infections in the urinary tracts, burn damage wounds, respiratory infections, and in lungs with cystic fibrosis (29). In this study, 48.3% of the chronic wounds were found to contain P. aeruginosa. Similarly, the highest prevalence was reported in hospitalized burn patients by Azzopardi et al. (30). Storm-Versloot et al. (31) also reported even a higher prevalence, with 52.2% of chronic wounds presenting this bacterium.
Biofilm production ability of microbial communities makes threats in case of wounds infection because immune system access to biofilm is very limited due to a thick layer. According to the current research, 79.5% of the P. aeruginosa isolates were found to be biofilm producers, whereas biofilm formation of P. aeruginosa was also found most prevalent in clinical isolates and had high associations in chronic wound infections (32). In our study, diabetic ulcers were found to be frequent, representing 43.96% of the chronic wounds. In diabetic ulcers, the patient’s condition as well as the environment may act together with other associated problems in the patient to affect the ecology of the wound (33). Diabetic patients always have decreased immune response with a lower resistance to microbial infections. Diabetic foot infection is the most common and severe complication in patients who suffer from diabetes mellitus. In this type of patients, wounds mostly are contaminated significantly by non-replicating bacteria (34). During the treatment of chronic wounds, different groups of antibiotics are used usually.
Treatment failure and increasing prevalence rate of infection in chronic wounds show the ability of microbial communities to resist the antibiotics. Based on the current findings, isolates from chronic wounds showed less resistance to amikacin and higher resistance to other antibiotics as shown in Figure 1. Isolates from chronic wounds showed the highest level of resistance to the antibiotic. Similarly, Moore and Flaws (35) reported that P. aeruginosa is highly resistant to antibiotics.
6. Conclusions
Due to the biofilm production ability of P. aeruginosa, it becomes more virulent and cannot eradicate easily from chronic wounds. Major reasons for antibiotic resistance are over use of antibiotics in daily life. This is one of the basic burdens on public health.