1. Background
Acinetobacter baumannii is a non-fermentative Gram-negative coccobacillus that is associated with a variety of serious infections (1, 2). It is now proposed that A. baumannii is one of the most common microorganism that can cause infectious diseases in hospitals throughout the world (3). This opportunistic pathogen causes infections that cannot be treated well because of its ability to survive against most classes of antimicrobial drugs (3). This pathogen has many kinds of innate and acquired mechanisms for antibiotic resistance such as change of target site, over production of efflux pumps, low permeability of outer membrane, and inactivation of enzymes like β-lactamases (4).
Β-lactamases, which are grouped into four classes including A, B, C and D according to amino acid sequence identities, are enzymes that degrade β- lactam antibiotics (5). Β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, inhibit peptidoglycan transpeptidases. Group “D” or carbapenem-hydrolyzing class D β- lactamases (CHDLs) are known as oxacillinases because of their ability to hydrolyze oxacillin (6). These enzymes are classified into four main OXA subfamilies: OXA-23-like, OXA-24/40-like, OXA-58-like, and OXA-51-like that the latest one is chromosomally located in all A. baumannii strains. Thus, it is an important genetic marker in the identification of these bacteria (7, 8). OXA types have a weak hydrolysis property but they are usually associated with genetic elements such as insertion sequences to increase the expression of carbapenemases and also mobilize them (7, 9).
The most percentage of these insertion sequences is ISAba1, although the other IS elements like ISAba2, are associated with resistance, too. When the IS element is located at the upstream of a number of OXA-type β-lactamases genes, such as bla OXA-23-like genes, bla OXA-51-like genes, and bla OXA-58-like genes, it increases their expression to a level that confers resistance to carbapenem (5, 10, 11). Therefore, determining the prevalence of IS as well as the OAXs genes in A. baumannii in different hospitals and medical centers is very important.
2. Objectives
To our knowledge, there are no data regarding the dissemination of these OXAs genes along with IS family, especially ISAba2, in A. baumannii from different hospitals in Tehran, Iran. Since IS elements are transferable and they can be transferred and increase the characteristic of antibiotic resistance to other bacteria, in this study we aimed to evaluate the antimicrobial activity of 14 antibiotics against clinical isolates of A. baumannii collected from five hospitals in Tehran, Iran. Furthermore, the distribution of four subgroups of OXA carbapenemases genes as well as the presence of the insertion sequences ISAba1 and ISAba2 were considered in A. baumannii.
3. Methods
3.1. Bacterial Isolates
In this experimental study, a total of 105 clinical isolates of A. baumannii were collected from patients in five different hospitals in Tehran between 2014 and 2015. Phenotypic identification of the isolates as A. baumannii was performed by using biochemical tests like growth at 42°C, culture on selective media, negative oxidase test, lack of lactose fermentation, etc. The PCR amplification and sequencing of blaoxa-51-like genes were used as the final confirmation of A. baumannii species.
3.2. Antimicrobial Susceptibility Test
A panel of antimicrobial susceptibility testing of the isolates to 14 agents was determined by Kirby-Bauer disc diffusion method on Muller- Hinton agar (Merck, Germany) according to the current clinical and laboratory standard institute (CLSI) guidelines (2015). The following antimicrobials were used in the susceptibility testing: Amikacin (AMK: 30 µg); Ceftazidime (CAZ: 30 µg); Cefepime (FEP: 30 µg); Cefotaxime (CTX: 30 µg); Ceftriaxone (CRO: 30 µg); Ciprofloxacin (CIP: 5 µg); Gentamicin (GEN: 10 µg); Imipenem (IMP: 10 µg); Meropenem (MEM: 10 µg); Piperacillin (PIP : 100 µg); Piperacillin + Tazobactam (TZP: 100 + 10 µg); Tigecycline (TGC: 15 µg); Minocycline (MCN: 30 µg); Trimethoprim + Sulfamethoxazole (SXT: 1.25 + 23.75 µg). All of these antibiotics were purchased from ROSCO, DK.
3.3. DNA Extraction
Genomic DNA was extracted by standard DNA extraction kit (Bioneer, Korea) according to the manufacturer's instructions. A total of 5 μL of DNA extract was used for each reaction.
3.4. Molecular Detection of OXA Carbapenemases and IS Elements and Nucleotide Sequencing
All isolates were subjected to PCR for the detection of four main groups of OXA carbapenemases genes (bla OXA-51-like, bla OXA-23-like, bla OXA-58-like and bla OXA-24-like) as well as ISAba1 and ISAba2 elements. All primers used in this study were listed in Table 1. The PCR mixture contained 1 µL (0.5 µg concentration) of extracted DNA, 1 µL (0.8 µM concentration) each of forward and reverse primers, 9.5 µL of water, and 12.5 µL of Master Mix (Ampliqon, DK). Amplifications were performed on thermocycler (Eppendorf, Germany). The DNA thermal cycler device (Eppendorf, Germany) was programmed in the following amplification conditions: after initial activation at 94°C for 5 minutes, 30 cycles, each for 45 seconds at 94°C (denaturation), 53 - 55°C (annealing), and 45 seconds at 72°C (extension) were performed. In contrast to other genes, the annealing time for ISAba2 was 48°C. The final cycle was followed by 72°C for 7 minutes (Table 2). Amplified PCR products were analyzed by 1.5% agarose gel at 100 V for 50 minutes in 1X TBE containing Red safe (INTRON). The PCR purification kit (Ampliqon, DK) was used to purify PCR products. The DM 100 - 2300 base pair ladder was used in this study (SMOBIO). Photography on gel was performed with UV doc (uvitec). PCR products were purified by using PCR purification kit (Bioneer Co., Korea), sequencing was performed by Bioneer Company (Korea), and their results were sent to gene bank.
| Primers | Nucleotide Sequences (5' to 3') | Amplicon Size, bp |
|---|---|---|
| OXA 51F | TAATGCTTTGATCGGCCTTG | 353 |
| OXA 51R | TGGATTGCACTTCATCTTGG | |
| OXA23F | GATCGGATTGGAGAACCAGA | 501 |
| OXA23R | ATTTCTGACCGCATTTCCAT | |
| OXA24F | GGTTAGTTGGCCCCCTTAAA | 246 |
| OXA24R | AGTTGAGCGAAAAGGGGATT | |
| OXA58F | AAGTATTGGGGCTTGTGCTG | 599 |
| OXA58R | CCCCTCTGCGCTCTACATAC | |
| ISAba1F | GTGCTTTGCGCTCATCATGC | 435 |
| ISAba1R | CATGTAAACCAATGCTCACC | |
| ISAba2F | AATCCGAGATAGAGCGGTTC | 1100 |
| ISAba2R | TGACACATAACCTAGTGCAC |
| Steps | OXA-51 | OXA-23 | OXA-24 | OXA-58 | ISAba1 | ISAba2 | Repeats |
|---|---|---|---|---|---|---|---|
| Activation | 94°C/5min | 94°C/5min | 94°C/5min | 94°C/5min | 94°C/5min | 94°C/5min | 1 cycle |
| Denaturation | 94°C/45s | 94°C/45s | 94°C/45s | 94°C/45s | 94°C/45s | 94°C/45s | 30 cycles |
| Annealing | 54°C | 53°C | 54°C | 54°C | 55°C | 48°C | |
| Extension | 72°C/45s | 72°C/45s | 72°C/45s | 72°C/45s | 72°C/45s | 72°C/45s | |
| Final extension | 72°C/7min | 72°C/7min | 72°C/7min | 72°C/7min | 72°C/7min | 72°C/7min | - |
4. Results
In this study, we investigated 105 clinical isolates of A. baumannii from five hospitals in Tehran. Following the conventional identification methods, the antimicrobial susceptibility testing was performed. The results of susceptibility testing are presented in Table 3. High levels of resistance shown in this table are phenotypes often observed in A. baumannii. Indeed, 100% of the isolates were resistant to Cefotaxime and resistance rates to the other cephalosporins were also very high. Carbapenems like Imipenem and Meropenem had also high rates of resistance. The lowest range of resistance was related to trimethoprim/sulfamethoxazole.
| Antibiotics | Concentrations, μg | Sensitive | Resistant |
|---|---|---|---|
| Minocycline | 30 | 7 (6.66) | 98 (93.33) |
| Trimethoprim/Sulfamethoxazole | 1.25 + 23.75 | 8 (7.61) | 97 (92.38) |
| Piperacillin | 100 | 2 (1.9) | 103 (98.09) |
| Ceftazidime | 30 | 4 (3.8) | 101 (96.19) |
| Cefotaxime | 30 | 0 (0) | 105 (100) |
| Gentamicin | 10 | 4 (3.8) | 101 (96.19) |
| Ciprofloxacin | 5 | 1 (0.95) | 104 (99.04) |
| Amikacin | 30 | 7 (6.66) | 98 (93.33) |
| Tigecycline | 15 | 2 (1.9) | 103 (98.09) |
| Imipenem | 10 | 5 (4.76) | 100 (95.23) |
| Cefepime | 30 | 1 (0.95) | 104 (99.04) |
| Ceftriaxone | 30 | 1 (0.95) | 104 (99.04) |
| Meropenem | 10 | 2 (1.9) | 103 (98.09) |
| Piperacillin/tazobactam | 100 + 10 | 4 (3.8) | 101 (96.19) |
aValues are expressed as No. (%).
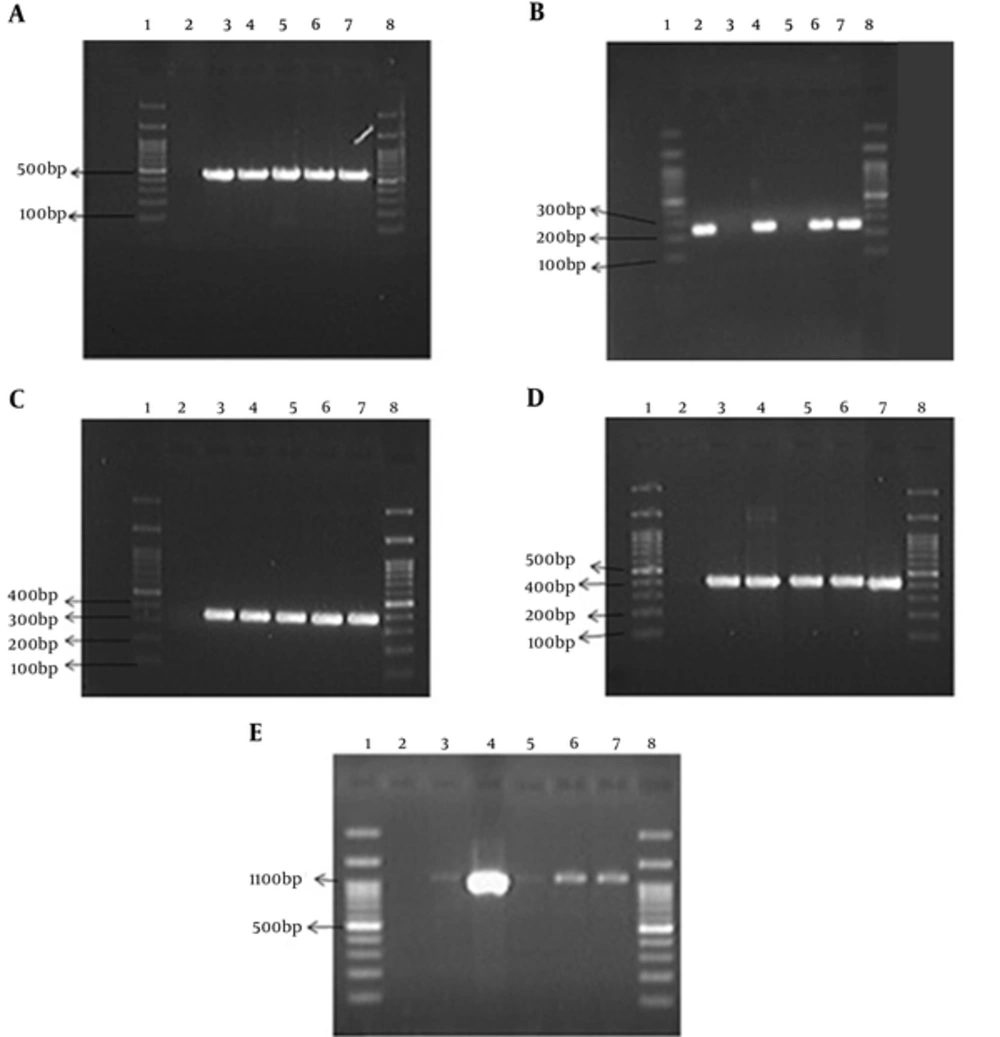
Distribution of OXA type genes and IS elements of A. baumannii isolated from hospitals in Tehran is shown in Table 4. All the isolates were positive for bla OXA-51-like genes that were confirmed to be A. baumannii and negative for carbapenemases belonging to OXA-58 family. Furthermore, 100% of the isolates had bla OXA-23-like genes that are one of the important resistance factors in this bacterium and 64.76% of the isolates had bla OXA-24-like genes. ISAba1 was found in all the analyzed strains of A. baumannii from the subjected isolates. ISAba2 was detected in 92.38% of the isolates. Furthermore, agarose gel electrophoresis of PCR products of all OXAs genes and IS elements is depicted in Figure 1. It is well elucidated that OXA 51 and ISAba1 are present in all samples, but we did not find OXA-58 in samples provided form the study hospitals. The presence of OXA- 23 and ISAba2 was confirmed in the most cases. The accession number for blaOXA-23 gene is KU310908.1.
| Genes | Percentage | |||||
|---|---|---|---|---|---|---|
| Total | Loghman | Motahari | Taleghani | Milad | Mofid | |
| OXA 51 | 105 (100) | 17 (100) | 33 (100) | 3 (100) | 48 (100) | 4 (100) |
| OXA23 | 103 (98.09) | 17 (100) | 31 (93.93) | 3 (100) | 48 (100) | 4 (100) |
| OXA58 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| OXA24 | 68 (64.76) | 17 (100) | 31 (93.93) | 3 (100) | 14 (11.53) | 3 (75) |
| ISAba1 | 105 (100) | 17 (100) | 33 (100) | 3 (100) | 48 (100) | 4 (100) |
| ISAba2 | 97 (92.38) | 12 (70.58) | 32 (96.96) | 3 (100) | 46 (100) | 4 (100) |
aValues are expressed as No. (%).
5. Discussion
Acinetobacter baumannii has emerged as one of the most troublesome pathogens for health care institutions globally (12). The ability of this microorganism to survive in both dry and wet conditions and their affinity to plastic and metal materials have lead to failure in hygiene prophylaxis. Changes in growth conditions may affect the transposition efficiency of several mobile elements and cause an increase in resistance by up-regulation or acquisition of different mechanisms and expression of resistance in plasmids and chromosomes particularly in presence of carbapenems (13). Carbapenems are the antibiotics of choice for treatment of infections caused by A. baumannii when these bacteria show resistance to other β-lactam antibiotics. However, resistance to carbapenems has increased and limited physicians in the choice of antibiotics. Generally, the carbapenem resistance is associated with the production of oxacillinase (OXA) enzymes. Nevertheless, metallo-β-lactamases can make resistance to carbapenems in A. baumannii (14, 15).
There are four main OXA subgroups associated with A. baumannii that their related genes are named: bla OXA-51-like, bla OXA -23- like, bla OXA -58- like and bla OXA -24 -like. The largest subgroup is OXA-51-like that corresponds to chromosome-encoded enzymes. With PCR performed in our study, all the 105 clinical isolates were positive for bla OXA-51-like gene. A survey by Visca et al. also demonstrated the possession of bla OXA-51-like gene among 117 strains of A. baumannii (16). Similar results were also obtained by previous studies (17-20). Our study confirmed that detection of bla OXA -51-like could be used as simple and reliable way to identify A. baumannii (18, 19, 21-23). The antimicrobial susceptibility profiles shown in Table 1 indicate that most of the 105 isolates showed high levels of resistance, a phenotype often observed in Acinetobacter.
All the isolates were resistant to Cefotaxime while resistance to other cephalosporins was also high. These results are in accordance with those of other studies (19, 20, 24, 25). Unfortunately, the resistance of A. baumannii was not only to β - lactams including penicillin, 3rd generation cephalosporin, and carbapenems, but also to other drug divisions, including aminoglycosides and fluoroquinolones (21). These high rates as shown in Table 1 were observed toward antibiotics such as Piperacillin, Piperacillin-tazobactam, Gentamicin, Minocycline, and Ciprofloxacin, too. In this study, the rates of resistance to imipenem and meropenem were 95.23% and 98.09%, respectively.
The results also showed that the prevalence rate of carbapenems and the frequency of multiple resistance Genes like OXAs are the causes of this problem. The blaOXA-23 carbapenemase gene has increasingly been reported worldwide (19). The OXA 23-β-lactamases were first identified in an A. baumannii isolate collected in Edinburg, United Kingdom. The resistance phenotype was transferable, indicating a plasmid location (10). OXA-23-like enzymes are able to hydrolyze oxyimino cephalosporins, aminopenicillins, piperacillin, oxacillin, and aztreonam in addition to the carbapenems (10). In this study, bla OXA-23-like gene was detected in 100% of the isolates, which is in accordance with other worldwide studies (17, 26-29).
Another gene encoding OXA-Type carbapenemase is bla OXA-24-like and its prevalence in isolates collected in the present study was 64.76%, most of which were found in isolates from one hospital. Distribution of bla OXA-24-like gene in Tehran was 15% in 2009 (30) and 17.3% in 2012 (31, 32). The last OXA-Type carbapenemase investigated in our study was bla OXA- 58- like gene. None of the one hundred and five clinical isolates in this study was positive for bla OXA- 58- like gene and these results are nearly similar to the findings of other studies in Iran obtained, for example, by Peerayeh et al. (30). The insertion sequences are frequently identified in association with an OXA β-lactamase gene (10). The most prevalent one of these insertion sequences is ISAba1. Insertion Sequence ISAba1, which has 11-bp inverted repeat sequences (IRs) flanked by 9-bp direct repeats of the target sequence, has been identified in A. baumannii. As one of the many IS elements, it contains promoters that play a role in the expression of antibiotic resistance genes (19).
In our study, all 105 clinical isolates were PCR positive for ISAba1 that is similar to previous studies (18). Turton and other authors have proposed that insertion of ISAba1 upstream of bla OXA- 51- like genes may provide the promoter to enhance gene expression potentially contributing to increased levels of resistance to carbapenems (17, 18). ISAba2 was the final genetic element we investigated. Similar to ISAba1, this element is likely to affect the expression of OXA carbapenemases. In the present study, the prevalence rate of ISAba2 was 92.38% (Table 3) and the high rate of this IS can explain the enhancement of promoters related to resistance genes.
6. Conclusions
In conclusion, it can be said that A. baumannii is an important pathogen in many countries. According to the results obtained in this study, it can be considered as red alarm microorganism in hospitals causing high rate of mortality and morbidity because it has multiple mechanisms for resistance and is not treated or treated hardly with common antibiotics. This microorganism especially in children and patients with immune deficiency can cause serious and longtime infections. Our study was done on some genetic elements that can have a major role in resistance to antimicrobial drugs especially the mobile elements that can be transferred within species to transform the antimicrobial pattern and cause an increase in resistance. Identification of these factors can help us control the infection and reduce the prevalence rate of this microorganism.