1. Background
Brucellosis is an infectious disease, caused by Brucella species and remains one of the most common zoonotic diseases worldwide. The disease is highly prevalent in many regions, particularly in central Asia, Middle East, the Mediterranean region, Africa, and Latin America (1, 2). Brucellosis causes abortion, sterility, mastitis in animals, and leads to great economic losses (3, 4). Transmission of brucellosis from animals to humans can occurr by consumption of infected unpasteurized milk and dairy products, inhalation of infected aerosolized particles, and through direct contact with infected animal parts (2, 3). Previously, despite the fact that brucellosis is considered to be an occupational disease, recently it has become an infectious disease affecting large populations in countries with poor nutritional hygiene and sanitation (2, 5). Among Brusella species, Brucella melitensis, B. abortus, B. suis, and B.canis are known to infect humans (5-7). In humans, brucellosis has a low mortality and high morbidity rate. It can be found as acute, subacute, localized, and chronic forms. Despite treatment, relapse rates of 6% to 11% can be observed (3, 4, 6, 8, 9).
There is no comprehensive data on the prevalence of brucellosis in children in Turkey. In the case series published in the recent years, brucellosis has been reported to cause arthritis, lymphadenopathy, splenomegaly, and maculopapular rash in children (9), and leads to relapses in some patients (6). Therefore, there is a need for molecular epidemiological studies that provide contributions to the control and treatment of disease in children. multiple locus variable number tandem repeat analysis (MLVA) typing is a useful method in outbreak and epidemiological trace - back investigations and in confirming laboratory or foodborne acquired infections (10, 11).
2. Objectives
In this study, it was aimed at determining species and biotypes of Brucella isolates that cause brucellosis in children, to determine antibiotic susceptibilities and to reveal genetic heterogeneity between Brucella isolates and transmission dynamics of isolates by the MLVA method.
3. Methods
3.1. Ethics Statement
The study was approved by the ethics and research committee of Yildirım Beyazit University (code: 26379996/148).
3.2. Brucella Isolates
The study was conducted on 77 Brucella isolates obtained from children hospitalized at the Diyarbakir Pediatric Disease Hospital, between 2000 and 2013. Patient demographic data including age, gender, symptoms, findings, and the seasonal distribution of the brucellosis were recorded. The isolates were cultivated on Brucella agar with hemin and vitamin K (HiMedia Laboratories) supplemented with 10% horse serum (Sigma - Aldrich) and Brucella selective supplement (HiMedia Laboratories). All isolates were identified and biotyped using conventional methods (requirement of CO2 for growth, urease activity, H2S production, sensitivity to the fuchsin and thionin dyes, lysis by Tbilisi phage, and agglutination with monospecific antiserum for A and M antigens), and tested for antibiotic susceptibility. Isolates were then taken to further molecular studies.
3.3. Antibiotic Susceptibility Tests
In vitro determination of antibiotic efficacy against Brucella strains is based on detection of minimal inhibitory concentration (MIC) values. For this purpose, in the current study, the E - test method was used as it is a reliable, reproducible, less labor-intensive, less time - consuming, and more practical micro - dilution method (12). To determine the MIC, 0.5 McFarland turbidity suspension of bacteria was spread on Mueller Hinton agar medium supplemented with 5% sheep and E - test strips (AB Biodisk, Solna, Sweden) for Gentamicin (GM), Rifampicin (RIF), and Doxycycline (DC), Tigecycline (TC), and Ceftriaxone (CRO), trimethoprim - Sulfamethoxazole (SXT) were placed on the agar plates (12, 13). The plates were incubated at 37 °C for 48 hours. The MIC was considered the value at which the inhibition zone intercepted the scale on the E - test strip. Brucella melitensis M16 strain (ATCC 23456) was used as the positive control strain. The MIC values of doxycycline, ciprofloxacin, TMP - SMX, and rifampicin for Brucella spp. were interpreted as recommended by the clinical and laboratory standards institute (CLSI) guidelines for slow - growing bacteria, such as Haemophilus spp. (12-14). Since the breakpoints value for tigecycline have not been determined by CLSI, ≤ 0.25 as sensitive was considered as interpretive MIC breakpoints for tigecycline (12, 15).
3.4. Molecular Studies
3.4.1. DNA Isolation
DNA isolation was done by the simple thermolysis procedure. For this, a loop of bacterial colony was suspended in 200 μL of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) solution. The bacterial suspension was heated at 100°C for 15 minutes and then centrifuged for 10 minutes at 13 000 × g. Two microliters of supernatant was used as the template in the polymerase chain reaction (PCR) procedures (11).
3.4.2. Determination of Brucella Species by Real Time Polymerase Chain Reaction (PCR)
Brucella species were determined by real - time multiplex PCR using bcsp31 (Brucella spp), BMEI1162 (B. melitensis), and alkB (B. abortus) primers, described previously (16). An amplification mixture was prepared in a total volume of 25 μL, including 12.5 μL of Thermo Scientific Maxima SYBR Green qPCR Master mix 2X (Thermo scientific, USA), 10 pmol of each primer shown in Table 1, 8 μL of water, and 2.5 μL of DNA. The cycling conditions were as follows, initial denaturation at 95 °C for 10 minutes, followed by 45 cycles of 95 °C for 15 seconds, 60 °C for 60 seconds and 72 °C for seconds. A final, 5 - minute elongation step was done at 70 °C. Amplification signals were collected during the annealing step. Melting curve analysis conditions were as follows: Ramp from 50 °C to 99 °C; increased by 1 °C each step; rest for 90 seconds of pre - melt conditioning on the first step. Rest for 5 seconds followed each step thereafter. Amplification results were evaluated by melting curve analysis.
aOligonucleotide sequence provided in 5′ to 3′ orientation, F: Forward, R: Reverse.
3.4.3. Multiple - Loci Variable Number Tandem Repeat Analysis Studies
In this step, 16 tandem repeat loci were amplified using Real - time PCRs. For this purpose, 5 different multiplex PCRs and one monoplex PCR were prepared in a manner that avoids overlapping of the resulting PCR fragments. The 16 loci tested in the study were as follows: Panel 1: Bruce 6, Bruce 8, Bruce 11, Bruce 12, Bruce 42, Bruce 43, Bruce 45, and Bruce 55; Panel 2A: Bruce 18, Bruce 19, and Bruce 20; Panel 2B: Bruce 4, Bruce 7, Bruce 9, Bruce 16, and Bruce 30. Standardization procedures were performed using a standard strain, B. melitensis M16 strain (ATCC 23456). Sequences and concentration of the primers used in each PCR mixture are listed in Table 2. As indicated previously, the forward primers were labeled with fluorescent dyes (14). The PCR mixes were prepared in a total volume of 25 μL with the Dream Tag Master Mix (2x) (ThermoFisher Scientific Inc., USA), appropriate concentrations of each primer, and 5 μL DNA of B. melitensis M16 (ATCC 23456) strain. Amplification procedures were performed using a thermal cycler (RG6000; Corbett Research). Amplification conditions were as follows, an initial denaturation step at 96 °C for 5 minutes, followed by 30 cycles of 96 °C for 30 seconds, 60 °C for 30 seconds, and 70 °C for 30 seconds, with a final extension step at 70 °C for 5 minutes.
| PCR Mixtures | Loci | Forward Primer (F) | Reverse Primer (R) | Concentration of each Primer (Nmol) f/r |
|---|---|---|---|---|
| 1 | Bruce21 | Cy5 - CTCATGCGCAACCAAAACA | GATCTCGTGGTCGATAATCTCATT | 10/10 |
| Bruce45 | Cy5.5 - ATCCTTGCCTCTCCCTACCAG | CGGGTAAATATCAATGGCTTGG | 10/10 | |
| Bruce11 | DyeD2 - CTGTTGATCTGACCTTGCAACC | CCAGACAACAACCTACGTCCTG | 20/20 | |
| 2 | Bruce04 | DyeD2 - CTGACGAAGGGAAGGCAATAAG | CGATCTGGAGATTATCGGGAAG | 20/20 |
| Bruce43 | Cy5 - TCTCAAGCCCGATATGGAGAAT | TATTTTCCGCCTGCCCATAAAC | 10/10 | |
| Bruce08 | Cy5.5 - ATTATTCGCAGGCTCGTGATTC | ACAGAAGGTTTTCCAGCTCGTC | 10/10 | |
| 3 | Bruce09 | DyeD2 - GCGGATTCGTTCTTCAGTTATC | GGGAGTATGTTTTGGTTGTACATAG | 10/10 |
| Bruce18 | Cy5.5 - TATGTTAGGGCAATAGGGCAGT | GATGGTTGAGAGCATTGTGAAG | 20/20 | |
| Bruce42 | Cy5 - CATCGCCTCAACTATACCGTCA | ACCGCAAAATTTACGCATCG | 10/10 | |
| 4 | Bruce16 | DyeD2 - ACGGGAGTTTTTGTTGCTCAAT | GGCCATGTTTCCGTTGATTTAT | 20/20 |
| Bruce07 | Cy5 - GCTGACGGGGAAGAACATCTAT | ACCCTTTTTCAGTCAAGGCAAA | 10/10 | |
| Bruce 12 | Cy5.5 - CGGTAAATCAATTGTCCCATGA | GCCCAAGTTCAACAGGAGTTTC | 10/10 | |
| 5 | Bruce06 | Cy5.5 - ATGGGATGTGGTAGGGTAATCG | GCGTGACAATCGACTTTTTGTC | 10/10 |
| Bruce55 | Cy5 - TCAGGCTGTTTCGTCATGTCTT | AATCTGGCGTTCGAGTTGTTCT | 10/10 | |
| Bruce30 | DyeD2 - TGACCGCAAAACCATATCCTTC | TATGTGCAGAGCTTCATGTTCG | 20/20 | |
| 6 | Bruce19 | DyeD2 - GACGACCCGGACCATGTCT | ACTTCACCGTAACGTCGTGGAT | 10/10 |
aOligonucleotide sequence provided in 5′ to 3′ orientation.
3.4.4. Capillary Electrophoresis and Fragment Analysis
To control amplification, 5 μL of the PCR products were electrophoresed in 2% agarose gel. Next, capillary electrophoresis was done on Beckman Coulter CEQ 8000 DNA Analysis System (Beckman Coulter, USA). For this procedure, 3 μL of the PCR product was diluted in H2O at 1/50. Twenty microliters of Sample Loading Solution (SLS) and 0.5 μL of "map marker 1000" were mixed. Furthermore, 3 to 5 μL of the diluted product was added to the mixture of SLS and map marker. Samples were loaded on the plate and a drop of mineral oil was dropped on them and the plate was placed on the Beckman Coulter CEQ 8000 DNA Analysis System. Then, capillary electrophoresis was performed using the following conditions: denaturation at 90 °C for 120 seconds, injection at 2.0 kV/20 seconds, and separation at 3.0 kV/3 hour (18). By the size of MapMarker, fragment sizes of each locus were defined and then analyzed (18).
3.4.5. Statistical Analysis
After capillary electrophoresis, the PCR product size was determined as "base pairs" (bp) and the MLVA - 16 values for each isolate were created. The fragment sizes were converted to tandem repeat unit numbers using a published allele numbering system (17). In the cluster analysis, unweighted pair group method with arithmetic mean (UPGMA) algorithm and categorical distance were used. The MLVA - 16 Orsay genotypes of B. melitentis isolates were compared with reference strains and prior isolates (17). The chi - square test was used to determine the relationship between the characteristics of the patients and the genotypes.
4. Results
4.1. Patient Characteristics
Of the 77 patients, 40 (55%) were male and 37 (45 %) were female, with ages ranging from 2 to 16 years old (mean of 9.13 ± 3.6 years). All patients had a history of recently consumed unpasteurized milk or dairy products. Abdominal pain was the most common complaint observed in 58% of patients followed by arthralgia (%56) and myalgia (40%). The most frequent clinical findings were fever, arthritis, hepatomegaly, and splenomegaly. Brucellosis in children was frequently observed in late spring and summer (May, June, and July).
4.2. Brucella Strains and Antibiotic Susceptibility Result
By using conventional and PCR methods, all isolates were determined as B. melitensis biovar 3. Two of the B. melitensis isolates were resistant to ceftriaxone, while all were susceptible to doxycycline, gentamycin and ciprofloxacin, and trimethoprim - sulfamethoxazole. Breakpoint values for tigecycline and rifampicin were found to be 0.016 to 0.23 and 0.38 to 1.5 μg/mL, respectively (Table 3).
| Antibiotics | MIC Range (µg/mL) | MIC 50 | MIC 90 | MIC of B. melitensis M16 Strain (ATCC 23456) (µg/ml) |
|---|---|---|---|---|
| Ceftriaxone (CRO) | 0.94 - 2.00 | 0.19 | 0.19 | 0.047 |
| Trimethoprim - sulfamethoxazolea (SXT) | 0.002 - 0.032 | 0.023 | 0.032 | 0.008 |
| Rifampicina (RD) | 0.5 - 1.00 | 1.0 | 1.0 | 1.5 |
| Doxycyclne (DO) | 0.016 - 0.064 | 0.032 | 0.047 | 0.094 |
| Gentamycin (GN) | 0.038 - 0.50 | 0.38 | 0.5 | 0.25 |
| Tigecyclinea (TGC) µg/mL | 0.016 - 0.064 | 0.047 | 0.064 | 0.032 |
aThe MIC values of the trimethoprim portion of the drug with a 1/9th percentile for trimethoprim - sulfamethoxazole were shown.
4.3. Multiple Locus Variable Number Tandem Repeat Analysis
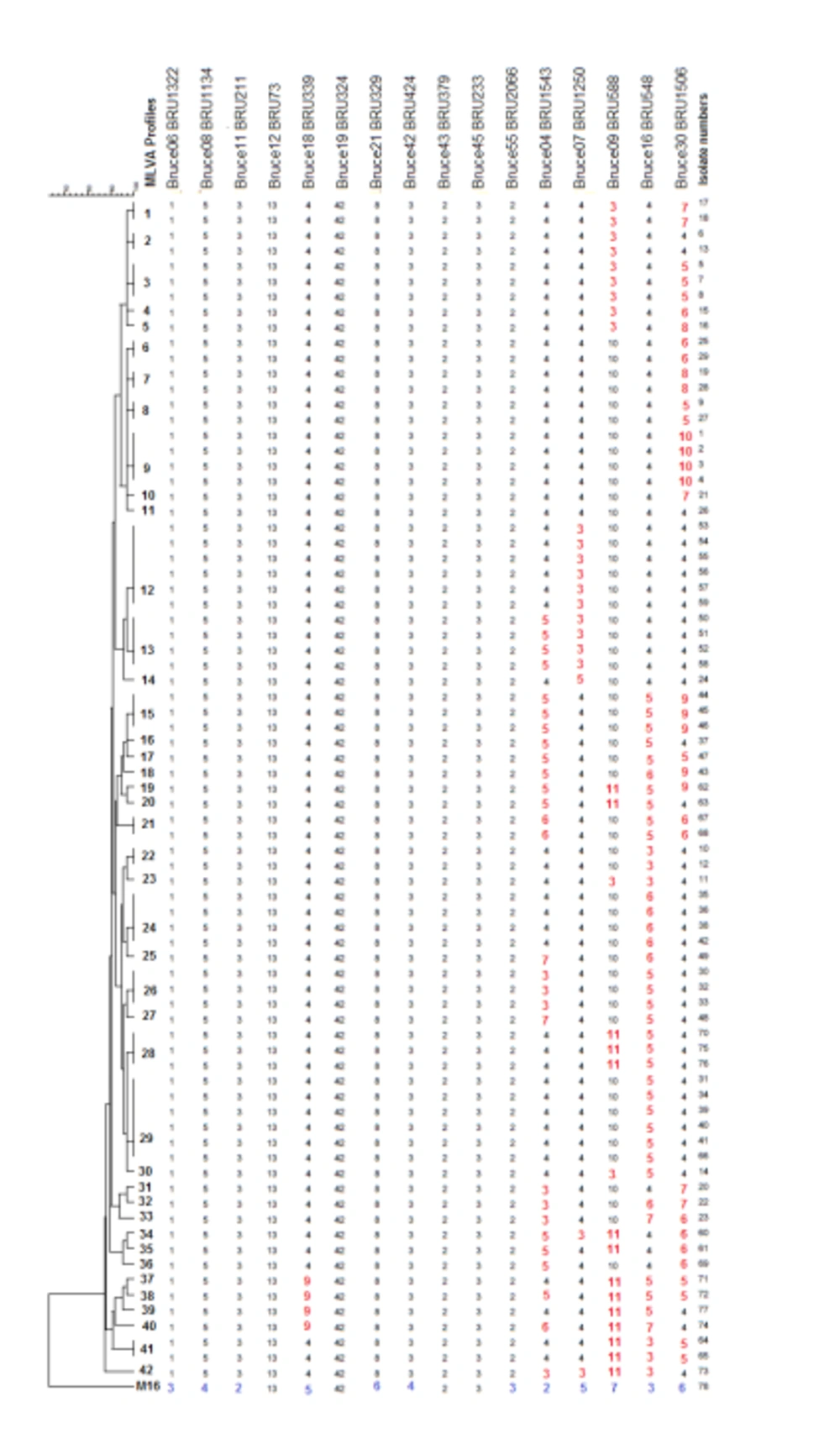
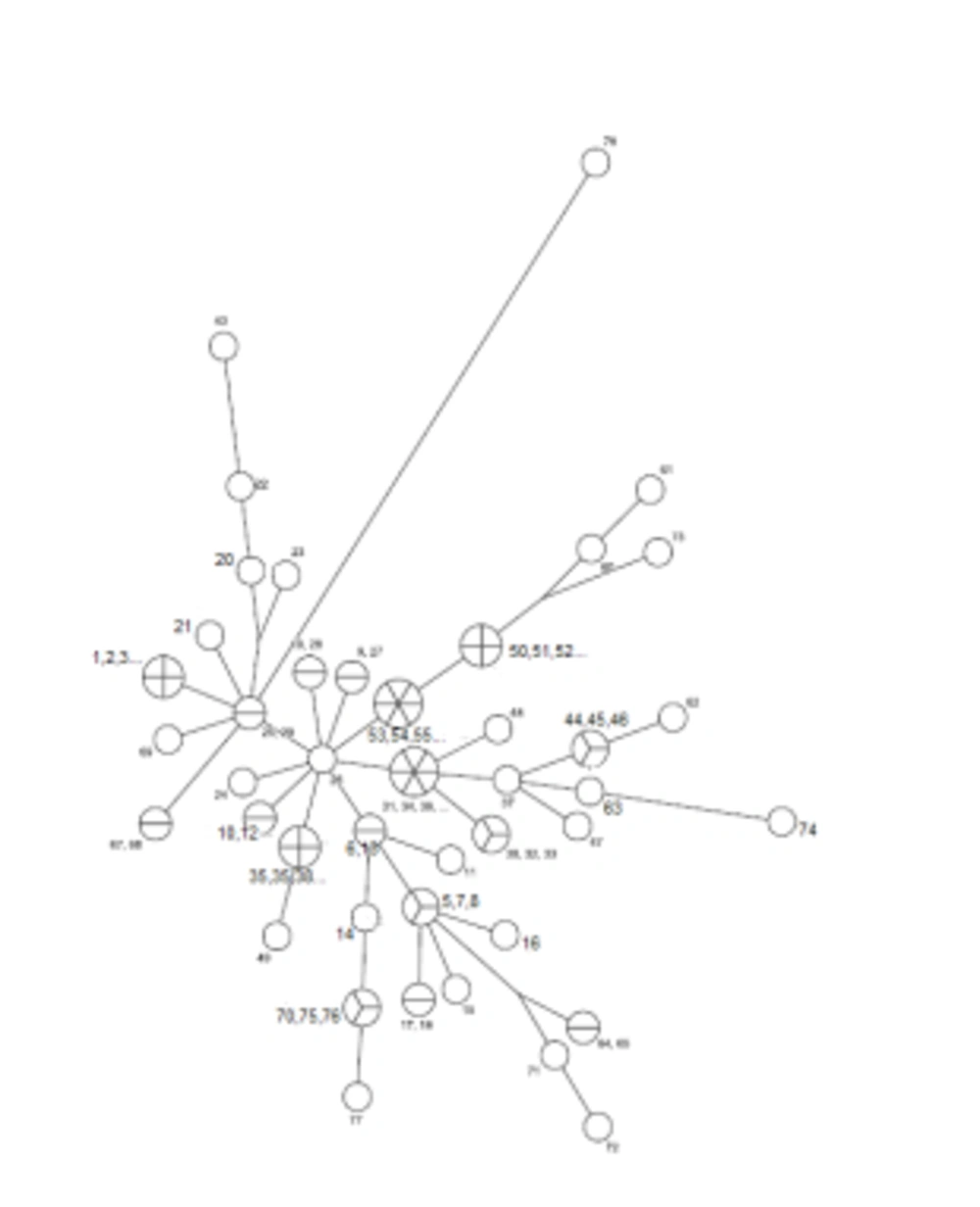
The MLVA - 16 Orsay genotyping analyzes revealed 42 different profiles in 77 isolates. Eighteen of these 42 genotyping profiles were clustered and each cluster contained 2 or more isolates. The clustering rate was 66.7%. The remaining 24 strains showed a specific profile. Single, double, and triple locus variants were detected in 32, 26, and 15 isolates, respectively. Four and five locus variants were observed in 3 and 1 isolate, respectively. The standard B. melitensis M16 strain (ATCC 23456) showed variations at 12 different loci. Bruce 30 displayed the highest discriminatory power, followed by Bruce 16, Bruce 9, Bruce 7, and Bruce 4, respectively. It was determined that the first 11 loci did not have discrimination power. According to MLVA 11, all strains were classified in genotype 122; according to MLVA8, all isolates were genotyped as 43. All of the isolates were in the Eastern Mediterranean genotype (Figures 1 and 2).
Cluster analysis of 77 Brucella isolates and B. melitensis M16 strain (ATCC 23456). Numbers on the branches indicated MLVA types defined in this study. The strains showing identical MLVA profiles were indicated with the same genotypes, such as isolate 35, 36, 38, and 42 in MLVA type 24, and isolate 30, 32, and 33 in MLVA type 26. Each unique profile indicated different numbers, such as isolate 15 in MLVA type 4. Numbers, which are below the loci indicate the number of tandem repeats. Loci that showed variation between tested isolates were indicated with red color. The variation detected in B. melitensis M16 (ATCC 23456) reference strain was labeled with blue color.
5. Discussion
Although there is no comprehensive data on the prevalence of brucellosis among children in Turkey, previous studies from the local areas indicate that brucellosis is still a major public health problem and causes difficulties and recurrences, especially in the treatment of child patients in this country. A previous study conducted on children in the capital city of Turkey showed that the most common complaints of children were fever (93.3%), malaise (86.6%), loss of appetite (80%), sweating (66.6%), and joint pain and/or swelling (53.3%); and the most frequent physical examination findings were arthritis (46.6%), lymphadenopathy (26.6%), splenomegaly (6.6%), and maculopapular rash (6.6%) (8). Relapse had been reported in one patient in that study. Another study involving 82 children with brucellosis in South - east Anatolia revealed that 76.8% of the cases were children of families working in agriculture and livestock farming. The reasons for admission to the hospital were fever (86.6%), joint pain (75.6%), fatigue (51.2%), and sweating (37.8%) (19).
The study performed on 32 children with brucellosis revealed that 87.5% of the cases had a history of eating raw milk and dairy products, and 12.5% had animal contact. In that study, relapse was seen in 3 patients, despite combined treatment (6). Similar to the literature data, all patients analyzed in this study had a history of recently consuming unpasteurized milk or dairy products, and abdominal pain, arthralgia and myalgia were the most frequent complaints and fever, arthritis, hepatomegaly and splenomegaly were the most frequent findings. From these data, it is suggested that parents of children must be trained about the use of pasteurized or boiled milk and avoid consuming cheese made from raw milk and raw meat. In addition to this, brucellosis should also be considered in children with abdominal pain and fever in areas where brucellosis is endemic.
Clinical studies demonstrated the need for molecular epidemiological studies that provide evidence - based data on cross - contamination, and source and spread of pathogens, which provide significant contributions to the control and treatment of disease. The MLVA method is a highly discriminating method that is frequently used in the genotyping of Brucella strains and allows comparison of results with worldwide data (20). In a study conducted in China, 12 B. melitensis isolates had been typed by MLVA 16. According to the results of MLVA - 8, these isolates were defined in 3 known genotypes, which were genotype 45, including 7 isolates, genotype 42 having 1 isolate, and genotype 62 with one isolate, as well as 2 new genotypes. According to MLVA 16 (Panel 1 + 2A + 2B), none of the isolates were found to be identical to the known genotypes (21). In a study from Kazakhstan, 128 B. melitensis strains were tested with MLVA. According to MLVA - 8 results, B. melitensis isolates were mostly found in genotype 42 (n = 108), followed by genotype 43 (n = 2), and 63 (n = 19) and these strains were related to the Eastern Mediterranean group (22).
The study from Italy identified 56 genotypes from 84 B. melitensis isolates, and it was reported that 81 isolates were associated with the Western Mediterranean group (10). In a study conducted on 75 strains of Brucella isolated in Kuwait, MLVA - 8 analysis identified all isolates as B. melitensis, and MLVA - 8, MLVA - 11 and MLVA - 16 typing divided the isolates to 10, 32 and 71 MLVA types, respectively. The combined minimum spanning tree analysis demonstrated that compared to MLVA types discovered all over the world, the Kuwaiti isolates were a distinct group of MLVA - 11 and MLVA - 16 types in the East Mediterranean Region (23). A study carried out by Kilic et al. that investigated the epidemiological relationship and genetic diversity among the 162 Brucella isolates collected from all geographical regions of Turkey in a period of 8 years showed that 161 isolates were identified as B. melitensis biovar 3. The MLVA - 16 typing resulted in 105 genotypes and high clustering rate was observed for half of the isolates and according to MLVA - 8, genotype 42 and 43 were recognized as the most common genotypes.
According to the results of this study, Brucella isolates were classified in the Eastern Mediterranean phylogenetic group and were associated with isolates of neighboring countries (11). In our original study that evaluated only childhood brucellosis, it was shown that transmission rate was very high (66.7%), most of the isolates were in Eastern Mediterranean phylogenetic group, and all of the isolates were in genotype 43. In the current study, greater discriminations was observed by MLVA panel 2 compared with panel 1. Similar to a previous study (11), Bruce 4, Bruce 16, and Bruce 30 loci were highly distinctive. From this result, it might be suggested that only Panel 2B loci can be used to define transmission dynamics of the strains in the low - income countries.
In children, combined treatment regimens of trimethoprim-sulfamethoxazole, rifampicin, and gentamycin were used in the treatment of Brucella infections, and doxycycline was added to therapy for children over 8 years of age (24). Identification of antibiotic susceptibility patterns of Brucella isolates is significant for determining appropriate treatment policies in regions where brucellosis is common due to treatment failures and relapse observations (11, 22-25). There was a limited number of studies on antibiotic susceptibilities of Brucella isolates recovered from children in Turkey. A study performed on brucellosis patients in a Van province from East Anatolia showed that doxycycline was the most effective antibiotic, followed by tigecycline, trimethoprim - sulfamethoxazole, and ciprofloxacin (22). In another study conducted on 56 Brucella isolates from the same province, MIC 90 values for doxycycline, streptomycin, rifampin, trimethoprim - sulfamethoxazole, and tigecycline were determined as 0.064 mg/L, 1 mg/L, 2 mg/L, 0.125 mg/L, and 0.094 mg/L for, respectively (26). In the current study, 2 of the 77 isolates were resistant to ceftriaxone, while all isolates were susceptible to doxycycline, streptomycin, and trimethoprim - sulfamethoxazole. Minimum inhibitory concentration intervals for tigecycline and rifampicin were 0.016 to 0.23 and 0.38 to 1.5 μg/mL, respectively. The data obtained from the results of antibiotic susceptibility showed that currently used antibiotic treatments regimens were valid, and tigecycline may be an alternative to treatment.
6. Conclusion
In conclusion, high clustering rate revealed that brucellosis among the children mainly results from common sources. Brucella isolates obtained from children in South east of Turkey belonged to B. melitensis biovar 3 and Eastern Mediterranean phylogenetic group. All of the isolates were defined as genotype 122, according to MLVA 11, and genotype 43, according to MLVA 8. Antibiotics currently in use for brucellosis treatment are still effective. In light of the current study, training activities were performed for public living in this region, including consuming pasteurized/boiled milk in children's diet, wearing appropriate clothes, face shield, and disposable gloves when working with animals and tissues. People under 18 years of age, pregnant females, and people, who are at higher risk for infection with weakened immune systems were recommended to be excluded from handling potentially infected animals or tissues. This study also demonstrated the necessity of controlling animal movements, and the sale of meat and dairy products, thus it was decided to share the study results with Republic of Turkey Ministry of Health.