1. Background
Cancer has been known as the leading cause of a variety of temporary or permanent physical and psychological changes in patients. Complications of cancer may lead to intolerable pain, hopelessness, fear, and even death. One of the most frequent cancers worldwide is leukemia, which occurred in 12.5 individuals per 100 000 people during 2011 (1). Infection is one of the most serious threats in patients with acute leukemia receiving chemotherapy and radiotherapy (2). Emergence of resistance to β-lactamse antimicrobial drugs began even before the first β-lactam penicillin was developed. The first β-lactamase enzyme was discovered in Escherichia coli prior to use of penicillin in clinical practice (3, 4).
The extensive use of β-lactams has led to the emergence of resistant isolates worldwide (5). Resistance to this class of antibiotics is mostly mediated by acquisition of β-lactamase genes located on mobile genetic elements, such as transposons or plasmids. Extended spectrum beta-lactamases (ESBLs) have increased among clinical isolates of members of the Enterobacteriaceae family during last four decades. High prevalence of infections caused by ESBL-producing Enterobacteriaceae at intensive care units (ICUs) was reported worldwide. Such drug-resistant bacterial infections are top concerns in people admitted to nursing homes, geriatric and oncology patients, transplant recipients, and pediatric populations. Failure to identify β-lactamase-producing isolates by laboratory susceptibility testing methods may lead to inappropriate empirical therapy and a subsequent increased mortality in these fragile and intolerable people (6).
Extended spectrum β-lactamase-producing Klebsiella pneumoniae and E. coli are progressively being identified in many parts of the world and are prevalent in countries located in the Asia-Pacific region (7). Over the past 20 years, CTX-M has emerged as a major type of ESBLs, which are now rapidly spreading among members of Enterobacteriaceae around the world (8). Since the discovery of the CTX-M-1 from a European patient in the late 1980s, more than 130 allelic variants of CTX-M were characterized (9). Among antimicrobial-resistant Gram-negative pathogens, organisms producing CTX-M-type ESBLs have become a serious threat to public health worldwide (10).
Insertion sequences (ISs) are the smallest transposable elements (< 2.5 kb) capable of independent transposition in an organism, thereby causing insertion mutations and genome rearrangements (11). Various elements may possibly be involved in the mobilization, spread, and expression of blaCTX-M genes, of which ISEcp1 is often associated with the upstream gene region. In addition, other insertion sequences, such as IS26, IS10, IS5, and IS903 have been detected surrounding the blaCTX-M resistance determinant (12).
2. Objectives
This study investigated the genetic environment of blaCTX-M genes in terms of the IS903, IS26, and ISEcp1 insertion sequences in E. coli and K. pneumoniae isolates recovered from patients with leukaemia in Tehran, capital of Iran.
3. Methods
3.1. Ethics Statement and Patient Consent
The present study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences with reference number IR.SBMU.SM.REC.1394.38. The authors confirm that all patients, and the patients’ parents provided their informed consent form to take part in the study.
3.2. Study Patients and Bacterial Isolation
Bacterial isolates included in this study were collected during May 2014 to October 2015 from two medical centers of Tehran, Iran: The Hematology-Oncology Research Center, Dr. Shariati Hospital (with a capacity of 857 bed), and the Mahak Pediatric Oncology Center, a 120-bed oncology hospital with an outpatient clinic. Sixty-one and nineteen subjects included in this study were patients with acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL), respectively. Bacterial isolates were recovered from various biological material, including blood, urine, wound, sputum, and vaginal swabs. All clinical specimens were immediately sent to the laboratory to be processed for microbial pathogens. Isolates were identified by using standard microbiological and biochemical methods (13). After identification, bacteria were stored in glycerol stock at -70°C before retrieval for testing.
3.3. Antimicrobial Susceptibility Testing
The susceptibility of clinical isolates to imipenem (10 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), levofloxacin (30 μg), amikacin (30 μg), ampicillin (30 μg), and gentamicin (10 μg) (Mast, UK) was evaluated by the Kirby-Bauer disk diffusion method, based on the Clinical Laboratory Standards Institute (CLSI) guidelines (14). The minimum inhibitory concentration (MIC) of selected antibiotics, including imipenem, ceftazidime, cefotaxime, and ciprofloxacin (Glaxo, UK) was determined by the reference broth microdilution method. All antibiotics were incorporated in Mueller-Hinton broth (Merck, Germany) in serial two-fold concentrations from 0.5 to 256 μg/mL. Quality control was done using the E. coli ATCC 25922 strain.
3.4. Screening Tests for ESBL Production
All isolates were evaluated for production of ESBLs by the phenotypic confirmatory disc diffusion method containing cefotaxime (CTX, 30 μg) and ceftazidime (CAZ, 30 μg) with CTX + clavulanic acid (CA, 10 μg) and CAZ + CA per disc (Mast Group, Merseyside, UK). The zones of inhibition were compared for the CTX and CAZ discs with that of the CAZ + CA and CTX + CA disc. An increase in zone diameter of ≥ 5 mm in the presence of CA was interpreted positive for the presence of ESBL in the test organism. Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as negative and positive controls for ESBL production, respectively (14).
3.5. PCR-Based Detection of blaCTX-M Type Genes and Sequencing Analysis
To determine the blaCTX-M variants, including blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, and blaCTX-M-25, PCR was performed on DNA extracted by a genomic DNA purification kit (Fermentas, Germany), using primers specific to each gene (Bioneer Company, Korea) (Table 1).
| Primer | Sequence (5’→3’) | Amplicon Size (bp) | Reference |
|---|---|---|---|
| CTX-M-1 | 781 | (15) | |
| F | CGTCACGCTGTTGTTAGGAA | ||
| R | ACGGCTTTCTGCCTTAGGTT | ||
| CTX-M-2 | 865 | (16) | |
| F | ATGATGACTCAGAGCATTCG | ||
| R | TGGTTACGATTTTCGCCGC | ||
| CTX-M-8 | 871 | (17) | |
| F | TGATGAGACATCGCGTTAAG | ||
| R | TAACCGTCGGTGACGATTTT | ||
| CTX-M-9 | 870 | (16) | |
| F | ATGTGACAAAGAGAGTGCA | ||
| R | CCCTTCGGCGATGATTCTC | ||
| CTX-M-25 | 924 | (18) | |
| F | CACACGAATTGAATGTTCAG | ||
| R | TCACTCCACATGGTGAGT | ||
| ISEcp1 | 526 | (19) | |
| F | GCAGGTCTTTTTCTGCTCC | ||
| R | TTTCCGCAGCACACCGTTTGC | ||
| IS903 | 588 | (20) | |
| F | CGTAGCGGGTTGTGTTTTCA | ||
| R | GGTAATTGATTCCACCGGGC | ||
| IS26 | 134 | (21) | |
| F | AGCGGTAAATCGTGGAGTGA | ||
| R | AGGCCGGCATTTTCAGCGTG |
Reactions were performed in a DNA thermal cycler (Eppendorf, Mastercycler gradient) in a 25-μL mixture containing 1 U of Taq polymerase (Bioneer Company, Korea) and 1× buffer consisting of 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, each dNTPs at a concentration of 250 μM, and primer at a concentration of 10 pmol. Thirty-six cycles were performed for each reaction, with the following temperature protocol for each cycle: 94°C for 60 seconds, 57°C for 60 seconds, and 72°C for 60 seconds. Sequencing of amplicons in both directions was done by the Bioneer Company (Daejeon, South Korea). The BLAST program from the national centre for biotechnology information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST) was used to analyse the nucleotide sequences.
3.6. PCR Assay for Insertion Sequences
All isolates were evaluated for the presence of three insertion elements ISEcp1, IS26, and IS903. The oligonucleotide primer sets specific for each IS are shown in Table 1. Sequencing was the same as described previously for CTX-M type genes.
3.7. Genetic Association of the CTX-M gene with ISEcp1, IS26, and IS903
The association of blaCTX-M to insertion sequences, such as ISEcp1, IS26, and IS903, was performed by PCR and sequencing. The reaction mixture and running protocols were the same as the above-described approach (12, 22).
4. Results
4.1. Isolation and Identification
A total of 56 E. coli and 24 K. pneumoniae isolates were recovered from urine (62.5%), blood (26.25%), sputum (8.75%), wound (1.25%), and vagina (1.25%). In total, 35% and 65% of the isolates were obtained from males and females, respectively. The distribution of patients’ age groups was as follows: ≤ 1 year, 3.75%; 2 to 17 years, 30%; 18 to 45 years, 40%; 46 to 64 years, 22.5%; and ≥ 65 years, 3.75%.
4.2. Antimicrobial Susceptibility Test and Phenotypic Confirmatory Disc
Both clinical isolates of E. coli and K. pneumoniae showed resistance towards ampicillin, while all K. pneumoniae isolates were susceptible to imipenem. Resistance rate of E. coli versus K. pneumoniae isolates to nine examined antibiotics was as follows: 44.64% versus 12.5% to ciprofloxacin, 7.14% versus 0% to imipenem, 30.35% versus 12.5% to gentamycin, 53.57% versus 16.66% to ceftazidime, 57.14% versus 20.83% to cefotaxime, 16.07 versus 8.33% to amikacin, 100% versus 100% to ampicillin, 62.5% versus 25% to levofloxacin, and 58.92% versus 20.83% to ceftriaxone (Table 2). Thirty-eight (67.85%) isolates of E. coli were capable of producing ESBL, with a variable range of resistance to all antibiotics tested from 13.15% against imipenem to 100% against ampicillin. In addition, 13 of 24 (54.16%) K. pneumoniae isolates exhibited ESBL-positivity, of which 100% and 100% were susceptible and resistant to imipenem and ampicillin, respectively. The MIC range, MIC50, and MIC90 determinations of four selected antibiotics against E. coli and K. pneumoniae along with the rate of resistant isolates are shown in Table 2. Of tested antibiotics, imipenem showed the lowest determinations of MIC50, and MIC90 (≤ 1 µg/mL) in both E. coli (MIC50 ≤ 1 µg/mL, MIC90 ≤ 1 µg/mL) and K. pneumoniae (MIC50 ≤ 1 µg/mL, MIC90 = 2 µg/mL) isolates (Table 2).
| Antimicrobial Agents | Bacterial Isolates | |||||
|---|---|---|---|---|---|---|
| MIC Range (µg/mL) | MIC50 | MIC90 | Antibiotic Susceptibility (%) | |||
| R | I | S | ||||
| E. coli (n = 56) | ||||||
| Ciprofloxacin | ≤ 2 - 256 | 4 | 8 | 44.64 | 8.92 | 46.44 |
| Cefotaxime | ≤ 2 - 256 | 16 | 64 | 57.14 | 3.57 | 39.29 |
| Ceftazidime | ≤ 2 - 256 | 16 | 64 | 53.57 | 5.35 | 41.09 |
| Imipenem | ≤ 1 - 8 | ≤ 1 | 2 | 7.14 | - | 92.86 |
| K. pneumoniae (n = 24) | ||||||
| Ciprofloxacin | 2 - 256 | 2 | 4 | 12.5 | - | 87.5 |
| Cefotaxime | ≤ 1 - 32 | ≤ 1 | 4 | 20.83 | - | 79.17 |
| Ceftazidime | ≤ 2 - 256 | ≤ 2 | 16 | 16.66 | 4.17 | 79.17 |
| Imipenem | - | ≤ 1 | ≤ 1 | 0 | - | 100 |
4.3. Detection of genes by PCR and Sequencing
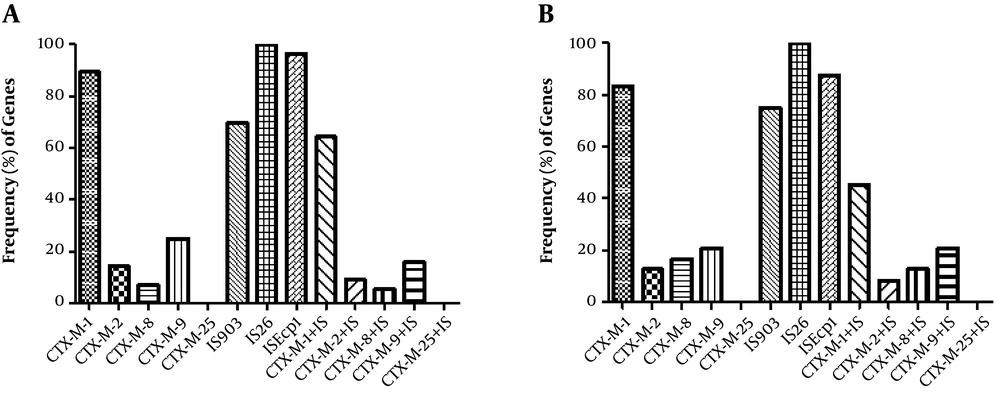
Molecular characterization of isolates revealed the presence of 89.28% (50/56) blaCTX-M-1, 14.28% (8/56) blaCTX-M-2, 7.14% (4/56) blaCTX-M-8, and 25% (14/56) blaCTX-M-9 in E. coli, while K. pneumoniae isolates were found to have 83.33% (20/24) blaCTX-M-1, 12.5% (3/24) blaCTX-M-2, 16.66% (4/24) blaCTX-M-8, and 20.83% (5/24) blaCTX-M-9. Prevalence of CTX-M variants among ESBL and non-ESBL-producing isolates and co-existence of resistance determinants is shown in Table 3. In addition, PCR identified the insertion sequences IS903 and ISEcp1 in 69.64% (39/56) and 96.42% (54/56) of E. coli isolates, respectively. Overall, 87.5% (21/24) and 75% (18/24) of K. pneumoniae were also positive for ISEcp1 and IS903 elements, respectively. The IS26 element was detected in all E. coli and K. pneumoniae isolates. Distribution of CTX-M variants and insertion elements among the isolates and co-existence of IS elements with CTX-M types is illustrated in Figure 1. Moreover, distribution of the insertion elements among the blaCTX-M-harboring ESBL and non-ESBL isolates is shown in Table 4. Sequencing analysis of the blaCTX-M-1 products revealed the presence of blaCTX-M-15 and blaCTX-M-55 variants among all clinical isolates.
| CTX-M Group | Bacterial Species, No. (%) | |||||
|---|---|---|---|---|---|---|
| Escherichia coli | Klebsiella pneumoniae | |||||
| ESBL-Positive (N = 38) | Non-ESBL (N = 18) | Total (N = 56) | ESBL-Positive (N = 13) | Non-ESBL (N = 11) | Total (N = 24) | |
| blaCTX-M-1 | 34 (89.47) | 16 (88.88) | 50 (89.28) | 11 (84.61) | 7 (63.63) | 20 (83.33) |
| blaCTX-M-1 + blaCTX-M-2 | 7 (18.42) | 1 (5.55) | 8 (14.28) | 1 (7.69) | 2 (18.18) | 3 (12.5) |
| blaCTX-M-1 + blaCTX-M-8 | 2 (5.26) | 1 (5.55) | 3 (5.35) | 1 (7.69) | 2 (18.18) | 3 (12.5) |
| blaCTX-M-1 + blaCTX-M-9 | 11 (28.94) | 3 (16.66) | 14 (25) | 1 (7.69) | 2 (18.18) | 4 (16.66) |
| blaCTX-M-1 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-2 | 7 (18.42) | 1 (5.55) | 8 (14.28) | 1 (7.69) | 2 (18.18) | 3 (12.5) |
| blaCTX-M-2 + blaCTX-M-8 | 1 (2.63) | 5.55 (1) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-2 + blaCTX-M-9 | 4 (10.52) | 5.55 (1) | 5 (8.92) | 0 (0) | 2 (18.18) | 2 (8.33) |
| blaCTX-M-2 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-8 | 2 (5.26) | 2 (11.11) | 4 (7.14) | 1 (7.69) | 0 (0) | 4 (16.66) |
| blaCTX-M-8 + blaCTX-M-9 | 1 (2.63) | 0 (0) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-8 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-9 | 14 (36.84) | 0 (0) | 14 (25) | 0 (0) | 3 (27.27) | 5 (20.83) |
| blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-1 + blaCTX-M-2 + blaCTX-M-8 | 1 (2.63) | 0 (0) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-1 + blaCTX-M-2 + blaCTX-M-9 | 4 (10.52) | 1 (5.55) | 5 (8.92) | 0 (0) | 2 (18.18) | 1 (4.16) |
| blaCTX-M-1 + blaCTX-M-2 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-1 + blaCTX-M-8 + blaCTX-M-9 | 1 (2.63) | 0 (0) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-1 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-2 + blaCTX-M-8 + blaCTX-M-9 | 2 (5.26) | 1 (5.55) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-2 + blaCTX-M-8 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-2 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-8 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-1 + blaCTX-M-2 + blaCTX-M-8 + blaCTX-M-9 | 1 (2.63) | 0 (0) | 1 (1.78) | 0 (0) | 1 (9.09) | 1 (4.16) |
| blaCTX-M-2 + blaCTX-M-8 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-1 + blaCTX-M-8 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| blaCTX-M-1 + blaCTX-M-2 + blaCTX-M-8 + blaCTX-M-9 + blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| CTX-M Group | Bacterial Isolates, No. (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli (N = 56) | Klebsiella pneumoniae (n = 24) | |||||||
| ISEcp1 | IS26 | IS903 | CTX-M + ISa | ISEcp1 | IS26 | IS903 | CTX-M + ISa | |
| blaCTX-M-1 | 50 (89.28) | 50 (89.28) | 37 (66.07) | 36.56 (64.28) | 14.24 (58.33) | 20 (83.33) | 14 (58.33) | 13 (45.16) |
| blaCTX-M-2 | 10 (17.85) | 8 (14.28) | 6 (10.71) | 5 (8.92) | 3 (12.5) | 3 (12.5) | 2 (8.33) | 2 (8.33) |
| blaCTX-M-8 | 3 (5.35) | 4 (7.14) | 4 (7.14) | 3 (5.35) | 4 (16.66) | 4 (16.66) | 3 (12.5) | 3 (12.5) |
| blaCTX-M-9 | 14 (25) | 14 (25) | 9 (16.07) | 9 (16.07) | 5 (20.83) | 5 (20.83) | 4 (20.83) | 4 (20.83) |
| blaCTX-M-25 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
a CTX-M +IS, coexistence of IS types with CTX-M types.
5. Discussion
Leukemia makes up about one-third of all malignancies in the 0- to 14-year-old age group. The most common subtype, acute lymphocytic leukemia (ALL), represents about 80% of these cases (22, 23). A total of 6000 new cases (3400 males and 2600 females) of aLL are diagnosed annually in the US. It occurs mainly in children and around 60% of cases are children and adolescents aged less than 20 years. Up to 90% of children affected by ALL survive, although it is necessary to improve the treatment of infants and adults (24). Severe, life-threatening infections continue to be a major cause of morbidity and mortality in patients with acute leukemia undergoing intensive chemotherapy (25). It has been reported that several risk factors, including neutropenia, decreased immunity due to underlying diseases, hemorrhagic diathesis of skin and mucosal tissue, severe mucositis, as well as central venous catheters (CVCs) increase the susceptibility to infections in leukemic patients (20, 26).
Specifically, multidrug-resistant organisms and even those previously considered harmless have currently emerged as a therapeutic challenge and can be catastrophic in this cohort of patients. Furthermore, ESBL-producing bacteria, such as Klebsiella spp. and E. coli, have become resistant to cephalosporins, fluoroquinolones, and penicillin classes of antibiotics, posing a threat to public health (27). This study focused on the detection of blaCTX-M genes, as an important mechanism of resistance to oxyimino cephalosporins (cefotaxime and ceftriaxone) in K. pneumoniae and E. coli, and analysis of the genetic organization of these determinants in terms of different insertion elements involved in their mobilization (7).
The high occurrence of ESBL-producing E. coli and K. pneumoniae isolates recovered from leukaemia patients admitted to therapeutic centres is alarming (28). Recently, Liu et al. reported even a higher prevalence (68.2%) of these isolates that were mainly positive for the blaCTX-M-15 (29). In a study from India, 48.27% of isolates were found to be ESBL-producers. Of these, 55.69% and 44.31% were identified as E. coli and K. pneumoniae, respectively (7). Yousefichaijan et al. showed that 53% of E. coli strains, isolated from infected children, were ESBL producing (30). In another study, Samet et al. showed that 0.41% of E. coli isolates from the leukemia patients were ESBL-producers (31). In the present study, blaCTX-M genes were detected in both clinical isolates of ESBL- and non-ESBL-producing E. coli and K. pneumoniae. CTX-M–type ESBL was first reported among E. coli isolates from Iran, during year 2008 (32). The percentage of E. coli and K. pneumoniae isolates carrying the blaCTX-M has increased significantly since then. In the previous studies during 2012 to 2013, blaCTX-M determinants was detected among 74% of ESBL-producing E. coli and the 58.33% of K. pneumoniae isolates (33, 34). To date, CTX-M–producing E. coli and K. pneumoniae were recognized in other parts of the country, including northern Iran (Enterobacteriaceae, 2014 to 2015, n = 8) (35), Kashan (Klebsiella spp, 2012 to 2013, n = 9) (36), Shiraz (2010, n = 2) (37), and Tehran (K. pneumoniae, 2015 to 2016, n = 40) (38).
The results revealed that the CTX-M-1 group, mainly CTX-M-15, was the most prevalent genotype, followed by CTX-M-9. The prevalence of CTX-M-15 in the current study was similar to that of previous reports. These findings also agree with recent studies from China and other countries, in which CTX-M-15 was reported as the dominant ESBL genotype (29). In the study by Chen et al., CTX-M-2 and CTX-M-8 groups of ESBLs were identified only in one isolate of K. pneumoniae (3). Xia and coworkers reported that 96.2% (1, 124/1, 168) of the isolates harboured blaCTX-M determinants, with a 40.7% (457/1, 124) and 48.7% (547/1, 124) blaCTX-M-1 and blaCTX-M-9-positive subgroups, respectively (39). Mirkalantari et al. showed that imipenem, with 84.7% susceptibility, was the most effective antibiotic against K. pneumoniae. Seventy (63.63%) isolates had ESBL-positive results and 42 (60 %) of them were positive for the CTX-M-1 gene (40).
Findings from previous studies suggest that more than one mobile element may be associated with different members of the CTX-M group (41). In the present study, ISEcp1, IS26, and IS903 elements were found to be associated with blaCTXM-1, blaCTX-M-2, blaCTX-M-8, and blaCTX-M-9 in a significant percentage of studied isolates (P ≤ 0.05). In addition, a high presence of IS26 and IS903 elements was detected in CTX-M-1-producing isolates. It is interesting to note that the presence of IS26 flanking ISEcp1 may affect the impact of this insertion sequence on the mobilization and expression of the blaCTX-M-15 gene. These elements could play an important role in the spread of such ESBLs. Thus, IS26, IS903, and ISEcp1 may be efficient tools for mobilization and expression of β-lactamase genes (42).
5.1. Conclusions
Multidrug-resistant Enterobacteriaceae that produce ESBLs, such as the CTX-M enzymes, have emerged both within the community and hospital settings as a serious issue. The rapid diagnosis of ESBL-producing isolates is helpful in selection of appropriate antimicrobials for therapy and prevention of the spread of these strains. Clinical microbiology laboratories should consider detection of ESBL producing K. pneumoniae and E. coli isolates important. It is recommended to routinely check all antibiotic resistant K. pneumoniae and E. coli isolates for ESBL production. This study indicates that the high prevalence of antibiotic resistance, IS and CTX-M-producing E. coli and K. pneumoniae isolates could be a major concern and highlights the need of infection control measures.