1. Background
Acinetobacter baumannii is a Gram-negative bacillus, pleomorphic but almost rod-shaped or coccobacillus, oxidase negative, catalase positive, non-motile, non-spore former bacterium and obligate aerobic (1, 2). This organism can be found widespread in nature and have been separated from water, soil and various samples of animal and human. Acinetobacter baumannii is able to adhere to the biotic and abiotic surfaces like the hospital equipment’s and can survive on these dry surfaces for long periods of time (2-5). The above mentioned properties of these species and the emerging antibiotic resistance for A. baumannii, could be related to biofilm formation. Outer membrane proteins (OMPs) are the plentiful surface proteins of this organism and have been exhibited to be necessary for its adherence (3-5).
Acinetobacter species (especially A. baumannii) are resistant to the most of the achievable antimicrobial agents and clinically useful antibiotics such as: aminoglycosides, fluoroquinolones, carbapenems and β-lactams (1, 2, 6, 7). Strains of Acinetobacter species. specifically Multi-drug and pan-drug resistant A. baumannii have been greatly separated from compromised patients with an underlying infectious including pneumonia (both hospital and community acquired), wound sepsis, urinary tract infections, septicemia and meningitis (6, 8-11).
The treatment of these infections has become greatly difficult due to the multi-drug, pan-drug and extreme-drug resistant (MDR, PDR and EDR) A. baumannii, an important cause of nosocomial infections (6, 9-15). The most important way for restriction of infectious diseases is vaccination. Vaccines have nearly or completely, eradicated both bacterial and viral infections, for examples, approximately complete eradication of poliomyelitis, elimination of smallpox and a diminution of more than 95% in the prevalence of diseases such as measles, pertussis, diphtheria, mumps, rubella and tetanus (16).
The second generation of vaccine can give some advantages such as complete prevention of infection and stability. In case of recombinant vaccines, gene(s) encoding antigenic proteins of a pathogen are inserted into the plasmid resulted in synthesis of these proteins inside the prokaryotic cells. After purification and injection of these proteins, followed by antigen presentation, an immune response can be induced (17). Therefore, we selected the high prevalence and conserved small outer-membrane lipoprotein A (SmpA) and used the pET expression system for gene expression and production of recombinant proteins in prokaryotic hosts (17).
It is necessary to mention that small outer-membrane lipoprotein A (SmpA, OmlA family) is associated with the YaeT complex. The YaeT complex is essential for the erection of OMPs into the outer membrane, and two of its members, YfiO and YaeT are necessary in Gram-negative bacterial in a way that deficiency of them may be incapable to comfortably assemble OMPs in the outer membrane. Strains with SmpA deficiency may present the heightened sensitivity to detergents and toxic miscellaneous toxic small molecules such as SDS, Deoxycholate, Tween-20, TritonX100, EDTA, Antibiotics, H2O2 and Paraquat (18, 19).
2. Objectives
The present study was aimed to perform the elementary steps in generation vaccine against MDR A. baumannii infections, including strains isolation from clinical samples in different wards, identification and evaluation of their antibiotic resistance patterns, cloning and expression of antigenic smpA gene A. baumannii in pET32(a). So, the results of present study will supply further experimental foundation for the development of A. baumannii vaccines.
3. Methods
3.1. Bacterial Isolates
Sixty-seven samples were collected from the different clinical infectious cases including wound pus (18 isolates), urinary tract infections (10 isolates), septicemia (14 blood isolates) and pneumonia (25 sputum isolates) in different wards of Kashani and Imam Ali hospitals Shahrekord, southwest Iran during September 2015 to April 2016. The majority of cases were hospitalized in the ICU. All the samples were cultured on solid media (McConkey; Merck, Germany) and blood agar. Afterwards A. baumannii were diagnosed with biochemical and bacteriological standard tests. The isolates were stored at -70°C in Trypticase Soy Broth (Merck, Germany) supplemented with 15% glycerol before being tested.
3.2. Antimicrobial Susceptibility Testing
The susceptibility of all antimicrobial agents’ isolates was determined by disk diffusion method, as recommended by the CLSI. The following antimicrobial agents (listed in Table 1) were provided by their manufacturers (MAST, Co, UK and Liofilichem, Co, Italy) for use in this study. The isolates were grown overnight on TSA supplemented with 5% sheep blood at 37°C. Acinetobacter baumannii ATCC19606 and E. coli ATTC25922 were taken from the IBRC, Iran, Karaj which was used for quality control purposes in antibacterial susceptibility determination.
| Antibiotic | S | I | R |
|---|---|---|---|
| Penicillin | |||
| Amoxicillin | 0 (0) | 0 (0) | 30 (100) |
| Cephalosporin | |||
| Cefoxitin | 2 (6.67) | 3 (10) | 25 (83.33) |
| Cefixime | 0 (0) | 0 (0) | 30 (100) |
| Cefotaxime | 0 (0) | 0 (0) | 30 (100) |
| Cefoprazone | 0 (0) | 0 (0) | 30 (100) |
| Carbapenms | |||
| Imipenem | 10 (33.33) | 3 (10) | 17 (56.67) |
| Doripenem | 4 (13.33) | 10 (33.33) | 16 (53.34) |
| Macrolide | |||
| Erythromycin | 0 (0) | 0 (0) | 30 (100) |
| Lincosamide | |||
| Clindamycin | 10 (33.33) | 12 (40) | 8 (26.67) |
| Quinolones | |||
| Nalidixic acid | 1 (3.33) | 5 (16.67) | 24 (80) |
| Ofloxacin | 1 (3.33) | 3 (10) | 26 (86.67) |
| Levofloxacin | 6 (20) | 4 (13.33) | 20 (66.67) |
| Sulfonamides | |||
| Sulfamethoxazole | 5 (16.67) | 3 (10) | 22 (73.33) |
| Tetracycline | |||
| Minocycline | 14 (46.67) | 10 (33.33) | 6 (20) |
| Glycylcycline | |||
| Tigecycline | 5 (16.67) | 7 (23.33) | 18 (60) |
| Polypeptides | |||
| Colistin | 23 (76.67) | 4 (13.33) | 3 (10) |
| Polymyxin B | 17 (56.67) | 8 (26.67) | 5 (16.67) |
In Vitro Susceptibilities for 30 Clinical Isolates of A. baumannii from Patients, in Different Wards of Imam Ali and Kashani Hospitals Determined by Disk Diffusion Methoda
3.3. DNA Extraction
Genomic DNA of A. baumannii strain was extracted by DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s guideline. Quality and quantity of the extracted genomic DNA were assessed by 1% agarose gel electrophoresis and the absorbance of purified DNA was measured at a wavelength of 260/280 nm by NanoDropND-1000spectrophotometer.
3.4. Gene Amplification
Gene encoding of the SmpA, nearly length in 417 bp was amplified by PCR method. Primers were designed with Gene Runner software according to the published sequence for smpA gene of the A. baumannii NCBI GenBank (Accession number: CP003967), and their exactitude was confirmed by BLAST (http//ncbi,nlm,nih.gov/BLAST). The sequences of these primers include smpA-Forward: 5'-ATAGTCGACATGCAAAAACTCGTGCTGAC-3' and smpA-Reverse: 5'-TCTGCGGCCGCTTATAGTGGTGGGGCAGTTA-3'. For subsequent cloning of the PCR-product fragments, SalI and NotI restriction sites were joined to the 5' end of these primers, respectively. The amplification was done using Thermal Cycler (Eppendrof, Germany), in a final reaction volume of 30 µL. The PCR mixture consisted of 4 µL of 10X PCR buffer, 2 µL MgCl2, 2 µL dNTPs (10 mM), 1.5 µL of each primer, 2 µL of DNA sample and 1 U of Taq DNA polymerase (Fermentas, Germany) and final volume was adjusted with nuclease-free water. The following conditions were applied: initial denaturation at 95°C for 5 minutes, followed by 32 cycles; denaturation at 94°C for 1 minute, annealing at 61°C for 1 minute, elongation at 72°C for 45 seconds. The program was ensued by final elongation at 72°C for 5 minutes and amplified sample was held at 15°C.
3.5. Valuation of Amplification Products and T/A Cloning
The PCR products were analyzed on 1.5% agarose gel electrophoresis, and photographed under UVIdoc gel documentation systems (Uvitec, UK). The amplification product was purified from the gel using gel extraction kit (Bioneer Co., Korea) according to the manufacturer protocol and cloned in pTZ57RTvector (Thermo Scientific, Lithuania). The engineered vector was transformed into the Escherichia coli Nova Blue competent cells by calcium chloride solution method using heat shock (at 42°C for 2 minutes and 4°C for 10 minutes). Transformed cells were cultured in LB agar (Merck, Germany) containing Ampicillin antibiotic (100 µg/mL), IPTG (0.1 M) and Xgal (20 mg/mL) for screening of recombinant vectors at 37°C overnight. The recombinant vectors were extracted from bacterial cells using plasmid mini extraction kit (Bioneer, Korea) using manufacturer’s guideline. PCR and double digestion with the SalI and NotI (buffer for double digestion: orange/1X) analysis were carried out for confirmation of cloning.
3.6. Sub-Cloning of smpA Gene Into pET32a (+)
After double digestion pTZ57R/T-smpA and pET32a (+) with the same restriction enzymes, smpA and linearized pET32a(+) were reclaimed by gel extract kit (Bioneer, Korea), and ligated by using T4 DNA ligase at 4°C overnight and generate the engineered pET32a-smpA according to directional cloning method. The E .coli Nova Blue was used for transformation by calcium chloride method and cultured in LB agar media containing 100 μg/mL ampicillin. This construct was confirmed by PCR, double digestion and sequencing.
3.7. Gene Expression
The pET32a-smpA was transformed into the competent expression host E.coliRosetta-gami2 (DE3) by calcium chloride method and plated onto the LB agar which contained 100 μg/mL of ampicillin and 35 μg/mL of chloramphenicol. The recombinant E. coliDE3 was grown in 5 mL LB broth containing ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL) on shaking incubator for overnight at 37°C. Then, 500 μL of culture was inoculated in 50 mL of LB broth medium at 37°C with agitation at 200 rpm. The cells grew until the optical density reached 0.6 - 0.8 at 600 nm. Expression of the SmpA protein was induced by the addition of 0.9 m M IPTG and incubated for five hours at 250 rpm/min, in 37°C. Afterwards, it was harvested by centrifugation and whole cell lysates was analyzed by SDS-PAGE 12%. In this case, E.coliDE3 without pET32a-smpA was used as a control. The E. coli DE3 was also transformed with empty vector PET32a which was induced by IPTG applying as a negative control. The mentioned control confirms that empty vector does not express any proteins with similar molecular weight to our target protein (SmpA).
3.8. RNA Extraction
Total RNA was extracted from 1 × 107 the recombinant E. coli DE3 containing PET32a-smpA induced by IPTG using RNX-Plus reagent (SinaClon, Iran) according to the manufacturer’s protocol. The concentration and purity of the genomic DNAs were evaluated by Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) reading at wave length 230 nm, 260 nm and 280 nm. The RNA was considered pure if the absorbance ratio of 260 nm/280 nm and 260 nm/230 nm was around 2.
3.9. Complementary DNA (cDNA) Synthesis and RT-PCR
Complementary DNA (cDNA) was synthesized using a reaction containing 500 ng/µL RNA, 100 mM dNTPs, 10 µL 5x RT-PCR buffer, 200 mM oligdT primer and 1 U/µL M-MuLV reverse transcriptase, (Invitrogen, USA) in a total volume 20 μL. The mixture was incubated at 42°C for one hour followed by enzyme inactivation; 15 minutes in 70°C. The smpA transcript was amplified using the specific primers as mentioned above. The reaction was performed in total volume of 20 μL containing 50 mM MgCl2, 0.25 mM of each dNTP, 2 pmol/L of each primer, 400 ng/ µL cDNA templates and1 U of Taq DNA polymerase (Fermentas, Germany). The final volume was adjusted with nuclease-free water. PCR was optimized in thermocycler and comprised of 5 minutes denaturation at 95°C for initial denaturation followed by 25 cycles of denaturation; 94°C for 1 minute, annealing; 61°C for 1 min, extension; 72°C for 45 seconds, final extension; 72°C for 2 minutes. A sample without DNA template (NTC) was included in all PCR reactions as negative control. The PCR products were then analyzed on 1% agarose gel electrophoresis dyed with 2% ethidium bromide and product bands were visualized under ultraviolet light.
4. Results
4.1. Bacterial Isolates and Antimicrobial Susceptibility Testing
In total 67 samples from patients aged between 20 days and 75 years, out of them, in 44.78% (n = 30) isolates were positive for A. baumannii. These bacteria were recovered from the wound pus (7 isolates), blood (6 isolates), urine (2 isolates) and sputum (15 isolates). Antibiotic susceptibility has been listed in High High resistance was observed among the tested antibiotics, excepted Colistin, Polymyxin B and Minocycline. One-hundred percent of the strains exhibited resistance to antibiotics Cefixime, Cefotaxime, Cefoprazone and Erythromycin. One hundred percent of these isolates at least were resistant to four generation of antibiotics and were denominated as MDR or EDR. Twenty resistance patterns of these mentioned isolates were determined by the same method and the results have been presented in Table 2.
| No. | Antibiotic Resistant Patterns | Frequency |
|---|---|---|
| 1 | AMX,CFX,TIG,CTX,CO,ERY,CC,NAL,CFM,IPM,PB,MIN,OFX,LOM,SMX,CFP,DP | 3 |
| 2 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,PB,OFX,LOM,SMX,CFP,DP | 1 |
| 3 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,MIN,LOM,SMX,CFP | 1 |
| 4 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,OFX,LOM,SMX,CFP,DP | 3 |
| 5 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,OFX,LOM,CFP,DP | 2 |
| 6 | AMX,CFX,TIG,CTX,ERY,CC,NAL,CFM,MIN,OFX,SMX,CFP | 1 |
| 7 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,PB,OFX,SMX,CFP | 1 |
| 8 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,OFX,LOM,CFP | 4 |
| 9 | AMX,CFX,CTX,ERY,NAL,CFM,IPM,OFX,LOM,CFP,DP | 1 |
| 10 | AMX,CFX,TIG,CTX,ERY,CFM,IPM,OFX,SMX,CFP,DP | 1 |
| 11 | AMX,CFX,TIG,CTX,ERY,CC,CFM,OFX,SMX,CFP,DP | 3 |
| 12 | AMX,CFX,CTX,ERY,NAL,CFM,OFX,LOM,SMX,CFP | 1 |
| 13 | AMX,CFX,TIG,CTX,ERY,NAL,CFM,IPM,SMX,CFP | 1 |
| 14 | AMX,CTX,ERY,NAL,CFM,IPM,OFX,LOM,CFP,DP | 1 |
| 15 | AMX,CTX,ERY,NAL,CFM,OFX,LOM,SMX,CFP,DP | 1 |
| 16 | AMX,CTX,ERY,CC,NAL,CFM,IPM,MIN,SMX,CFP | 1 |
| 17 | AMX,CFX,CTX,ERY,CFM,OFX,LOM,SMX,CFP | 1 |
| 18 | AMX,CTX,ERY,NAL,CFM,OFX,LOM,SMX,CFP | 1 |
| 19 | AMX,CTX,ERY,NAL,CFM,OFX,LOM,CFP,DP | 1 |
| 20 | AMX,CFX,TIG,CTX,ERY,CFM,OFX,SMX,CFP | 1 |
| Total (%) | 30 (100%) |
Antibiotic Resistance Patterns of A. baumannii Clinical Isolates from Patients with Nosocomial Infections, in Different Wards of Imam Ali and Kashani Hospitals
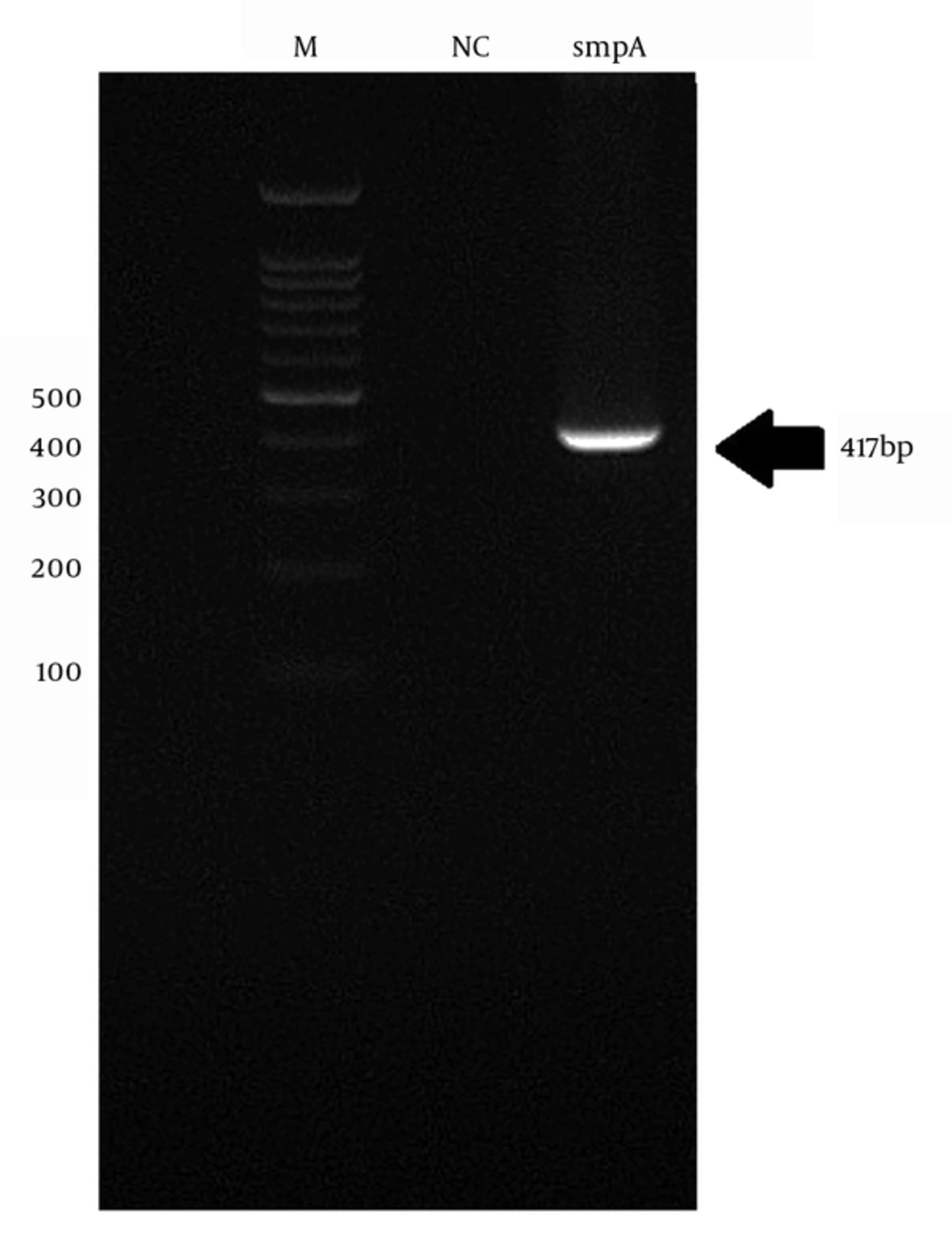
4.2. DNA Extraction and Amplification of smpA Gene
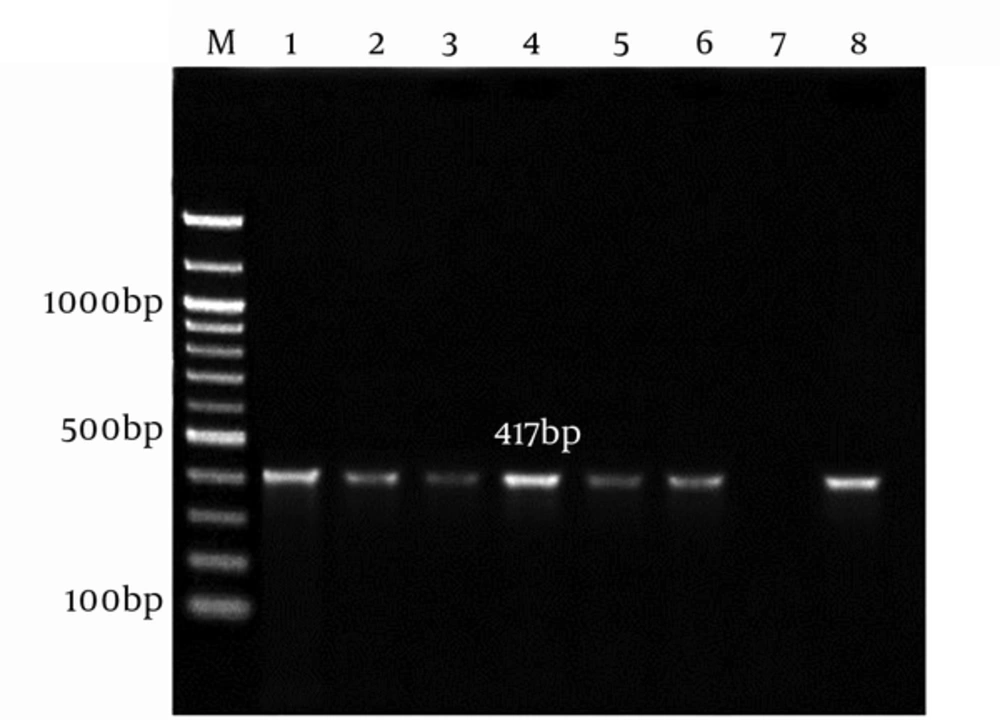
The genomic DNA of A. baumannii was successfully extracted through the amplification of smpA gene. The 417 bp PCR product was harmonized to the expected length (Figure 1).
4.3. T/A Cloning
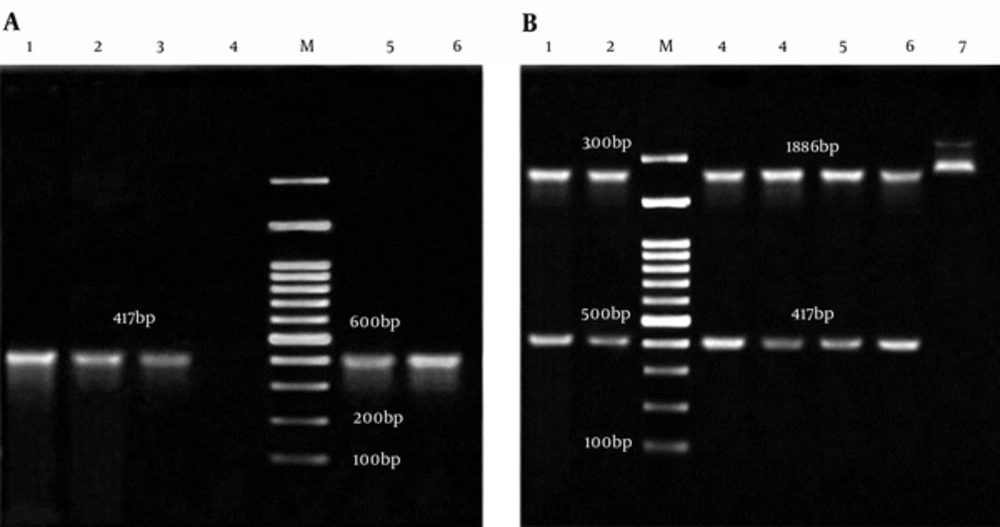
The appearance of white and blue colonies on LB agar plates was an evidence of successful transformation. Colony PCR and double digestion of pTZ57RT-smpA showed that smpA gene was cloned in pTZ57RT vector successfully (Figure 2A and 2B, respectively).
A, Colony PCR of smpA; Lane M: 100bp DNA marker; Lane 1 - 3 and 5, 6: an Amplification of Complete smpA gene (417 bp) from pTZ57RT-smpA; Lane 4: Negative Control; B, Analysis of pTZ57RT-smpA Double Digested; Lane M: 100 bp DNA Marker; Lane 1, 2 and 3 - 6: 2886 bp Segment pTZ57RT and Fragment 417 bp smpA; Lane 7: pTZ57RT-smpA undigested.
4.4. Sub-Cloning smpA and Sequencing
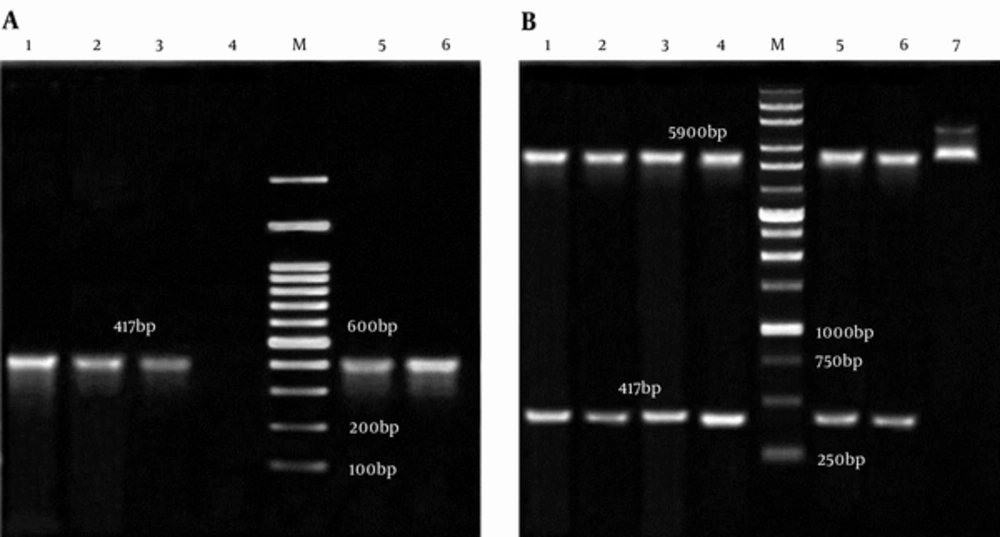
Analyzing of the double digested and colony PCR from pET32a-smpA showed that the engineered vector contained the gene of interest (Figure 3A and 3B, respectively). The sequencing of smpA gene showed more than 95% similarity with smpA published in the NCBI gene bank (Supplementary file 1).
A, Colony PCR of smpA; Lane M, 100 bp DNA Marker; Lane 1 - 3,5 and 6, an Amplification of Complete smpA Gene (417 bp) from pET32a –smpA; Lane 4, Negative Control; B, pET32a -smpA Double Digested; Lane M: 1 Kb DNA Marker; Lane 1 - 4,5 and 6, 5900 bp Segment pET32a and Fragment 417 bp smpA; Lane 7, pET32a-smpA Undigested.
4.5. Expression of the Recombinant Protein (rSmpA)
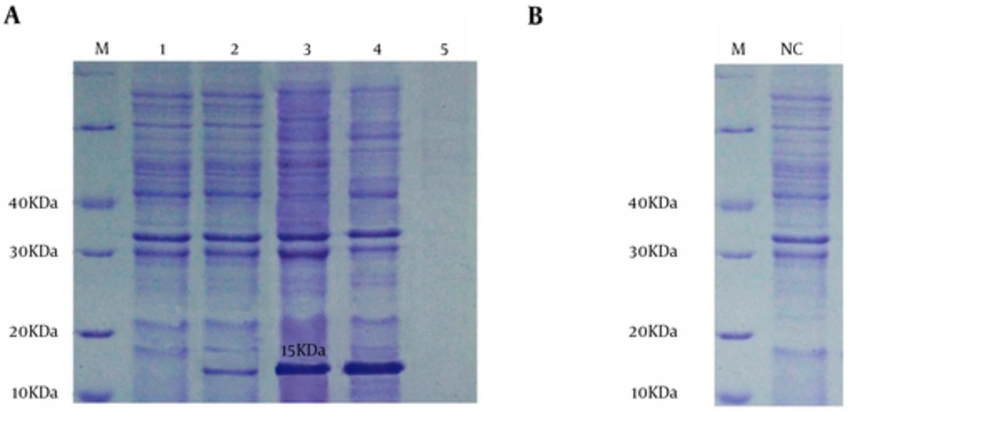
The pET32a-smpA in E.coli DE3 system was induced by IPTG at concentration of 0.9 mmol/L. The results of SDS-PAGE demonstrated that the pET32a-smpA efficiently produce rSmpA. The protein band in the range of molecular weight ~15 KDa was found in SDS-PAGE (Figure 4A). Addionally, to examine if the empty vector does not express any proteins with similar molecular weight to our target protein (SmpA), E. coli DE3 was also transformed with empty vector PET32a which was induced by IPTG. Our results showed that the empty vector does not produce any proteins with similar molecular weight to the SampA (Figure 4B).
A, lane M, molecular mass marker in 10 kDa; lane 1, whole bacterial lysate of the E. coli Rosetta-gami without pET32a-smpA; lane 2, the E. coliDE3 including pET32a-smpA was no induced; lane 3 - 4, the whole bacterial lysate of the E.coliDE3 contained recombinant pET32a-smpA was induced; lane 5, Blank control; B, IPTG-induced E. coli Rosetta-gami with emty pET32a vector.
4.6. The Expression Level Analysis of smpA gene in Recombinant Bacteria
The RNA was extracted from the recombinant E. coli DE3 containing PET32a-smpA induced by IPTG. The cDNA was then synthesized from the transcripts to amplify in RT-PCR using specific primers for smpA. The presence of 417 bp band was surfed by means of electrophoresis (Figure 5).
5. Discussion
Acinetobacter baumannii accounts as an important cause of endemic and sporadic nosocomial infections. Recently progressive significant increasing of antibiotic resistance in A. baumannii isolates, created a serious threat to hospitalized patients (6, 8-11). In our study, in around 45% of isolates A. baumannii was diagnosed, the majority of studied patients were hospitalized in the ICU and most samples were isolated from the sputum cultures (50%). According to Table 1, among the A. baumannii isolates, except against Colistin, Polymyxin B, Minocycline and Clindamycin, high rates of antibiotic resistance were seen; 100% were resistance to Cephalosporins and other antibiotics including Amoxicillin and Erythromycin; but the highest resistance to Carbapenms, Quinolones, Sulfonamides and Glycylcyclines were observed. These data are similar to recent researches reported that nosocomial infections caused by the Acinetobacter species are progressively becoming resistant to a broad spectrum of antibiotics (2, 8). In studies by Japoni-Nejad and Vakiliin on Iranians resistance to Imipenem and Meropenem, they found higher resistance rates than the observed data in our study (6, 8).
Multi drug resistant A. baumannii is a common challenge in treatment of hospital infections in many countries, including America, Italy, Korea, Greece, Belgium and Iraq (9-14).The spread on antibiotic resistance among Acintobacter species has emerged as an important challenge for Iranian infection diseases specialists and medical society too. A lot of MDR A. baumannii outbreaks have been announced in Iran (1, 2, 6, 20-22). An Iranian surveillance report of antibiotic resistance in 2001 - 2007 found that 40.6% of MDR A. baumannii isolates collected from CMC and local hospitals were Ampicillin and Sulfumetoxazole resistant (15). Another study in Tehran showed that 90% of A. baumannii isolates were resistance to Cephalosporins (1). In 2010, the Iran Surveillance demonstrated that the prevalence of A. baumannii antibiotic susceptibility to Gentamycin, Tobramycin, Kanamycin and Amikacin decreased gradually from 50%, 62.5%, 50% and 81.2% in 2002 to 28.1%, 25%, 15.6% and 25% in 2007, respectively.
Soroush reported that susceptibility rates to Cefteriaxon, Cefixime, Ceftizoxime, Cephalotinand Ceftazidime were 43.7%, 50%, 31.2%, 31.2% and 50% in 2001 decreased to 9.3%, 0%, 18.7%, 0%, and 0% in 2007 respectively (15); and A. baumannii Multi-drug and Extreme-drug resistant increased from 40.6% in 2002 to 95% in 2014 (8, 15). In addition, the distribution of healthcare-associated infections caused by A. baumannii resistant to Carbapenem in Iran has been significantly increased, and the frequency resistance rate to Carbapenem was increased progressively from 13.6% in 2008 (23) to the 83% in 2014 (8).
Our study showed that out of seventeen antibiotics, Colistin, Polymyxin B and Minocycline were suitable antibiotics to be used in infection medication, by considering of their side effects. However, the rest of the studied antibiotics were inefficient. Carbapenems have been reported operative antibiotic with lower resistance rate in some studies in Iran (20, 21), but our finding do not confirm this conclusion.
In a recent study in Iran, Imipenem resistance in enteric bacteria isolated from hospital infections have been reported equal to 33%, which is dissimilar with our results (22). We found that Meropenem and Imipenem were inefficient combat A. baumanni isolates by disc diffusion method. The findings of several studies have demonstrated that Colistin medication in patients with MDR A. baumanii caused an admirable clinical effect and little mortality (8, 24).Therefore, this increased usage perchance lead to Colistin resistance and choice of suitable antibiotic therapy is necessary. Different studies reported that Colistin resistant A. baumannii have increased gradually all over the world, including Korea (from 9.1% in 2002 to 30.6% in 2007) (25, 26), Spain (from 19.1% in 2005 to 40.7% in 2009) (27, 28) and lower frequency of resistance were observed in USA and South America (2.1% and 7.1 %, respectively) (26).
In the current study, a total 20 phenotypic patterns were observed, three strain resistance to 17 antibiotic and 100% isolates resistance to at least 5 antibiotics. Different patterns have been explained in earlier studies (6, 28) and evidently variable of resistance patterns in different populations might be due to some factors including: hospitalized patients staying, doctors, prescribing clothing, patients visiting, and expenditure policies (26-29), as well as differences in climatic condition and methods of sample collection. Increase of MDR and PDR organisms has led to anxiety in the medical society, hence, there is an urgent need to novel strategies to combat this MDR and PDR pathogen. Therefore, based on the urgent need for prevention of A. baumannii infections, we generated a recombinant pET-32a/smpA expression vector for produce rSmpA as a recombinant vaccine candidate. In order to achieve this goal, at first, smpA gene of A. baumannii was amplified and were cloned successfully in pTZ57RT vector and then sub-cloned in linearized pET32a expression vector and successfully pET-32a/smpA construct is generated. Based on our knowledge, this is the first report on the isolation, cloning and prokaryotic expression antigenic smpA gene detected the clinical strains of A. baumanni in the world.
Outer membrane proteins have critical functions in bacterial survival, host invasion, virulence, immune evasion, cell adhesion and cell death. Outer membrane proteins are mostly immunogenic and appearance on the cell surface, and seem to be potential antigen candidates. Several previous studies demonstrated that some protein components of A. baumannii outer membrane proteins and outer membrane vesicles were exceedingly immunogenic (3-5), so they were considered as potential antigen candidates for further recognition. Based on these findings, using information of reported clinical A. baumannii isolates, bioinformatics analyses on sequence conservation and possible classification of candidate proteins, the extremely conserved, secretory proteins or predicted membrane were chosen preferentially.
The SmpA is a candidate protein confirmed by bioinformatics estimation of A. baumannii proteome (30). It is a putative outer membrane assembly protein. Although its accurate role in virulence is unidentified, presence of SmpA in infective clinical isolates and in the outer membrane vesicles of infesting clinical strains makes it as a critical antigen. Bioinformatics analysis demonstrated its localization in outer membrane, no transmembrane helices, high adhesion probability, conservation between the different strains of A. baumannii (5, 18, 19). SmpA dissimilarity with mouse and human proteome indicated that the risk for SmpA-stimulated antibodies to cross-recognize human proteins is theoretically imperceptible, so this property presenting it as a highly potential candidate vaccine. Therefore produced SmpA in the present study is a potent antigen candidate for developing an effective vaccine for prevention of A. baumannii infections.
6. Conclusions
Result of this study indicated 100% of the investigated Acinetobacter isolates were tremendously resistant to Cephalosporins and high antibiotic resistance rates to other antibiotics such as Quinolones, Sulfonamides, Glycylcyclines and Carbapenms were also seen, that it may be due to unwise and inappropriate use of antibiotics to treat infections. The four most effective antibiotics against Acinetobacter were found to be Colistin, Polymyxin B, Minocycline and Clindamycin. Also, the recombinant form of the SmpA protein showed high expression in E. coli, so we suggested that this rSmpA protein may be suitable as a vaccine candidate. However, for evaluation such potential, further studies such as evaluation in animal models and Human clinical trials will be needed.