1. Background
Intestinal parasites are considered as the most common and serious public health problem particularly in low-income developing countries (1-7). Cryptosporidium is considered as an obligate intracellular protozoan parasite, which causes respiratory or intestinal disorders in wide range of vertebrates such as humans, rodents, livestock, etc. Oocysts, as an infective stage of the Cryptosporidium life cycle, are shed via feces of afflicted hosts into the environment and remain infective for a long time due to a high resistance to environmental conditions (8).
Cryptosporidiosis in immunocompetent individuals is mostly asymptomatic, although in immunocompromised patients, it causes a life-threatening and severe infection (8, 9). Cattle are the major hosts for several species of Cryptosporidium. Furthermore, cattle play a key role in maintaining the Cryptosporidium transmission cycle in surrounding regions. The parasite causes a neonatal illness in cattle, delayed growth, and weight loss, which imposes significant economic wastage on society (10, 11).
The diagnosis of Cryptosporidium spp. is routinely based on a direct observation of oocysts in stool samples under light microscope. Unfortunately, this method has a low sensitivity and is also not appropriate for determination of different species of Cryptosporidium (8). Molecular based techniques such as polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) have become popular worldwide (12, 13) due to high sensitivity and specificity in differentiate between Cryptosporidium species (7, 14-17).
2. Objectives
Lack of studies regarding the cryptosopridian infection in cattle of Ahvaz city, southwest of Iran, encouraged us to design present survey to determine the prevalence and molecular characterization of Cryptosporidium spp. using Nested-PCR-RFLP of small subunit-rRNA (SSU 18s rRNA) gene.
3. Methods
3.1. Study Area
Ahvaz city, capital of Khuzestan province, which is located in the southwest of Iran (31°50’N and 49°11’E), is ranked as the 7th largest city throughout the country and based on the latest census, its population is calculated about 1395184 and 352128 families. Weather temperature is highly variable throughout the year so that in the summer the temperature exceeds 50°C whereas in winter it falls to 5°C. Furthermore, the annual average rainfall is approximately 230 mm (7).
3.2. Sample Collecting and Processing
Initially, the city of Ahvaz was divided into 5 geographical locations (west, east, north, south, and center). A total of 240 cattle stool samples were collected from both urban and rural areas of the city of Ahvaz. Stool specimens (40 - 50 gr) were taken in labeled plastic containers and immediately transferred to the department of medical parasitology (Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran) for further testing. A questionnaire including some demographic information was filled out for each cattle. All stool samples were examined using optical microscope (× 1000 magnification) in order to detect the oocysts after concentration by sugar flotation method and staining with modified Ziehl-Neelsen.
3.3. DNA Extraction and Nested-PCR
The DNA extraction kit (QIAamp® DNA Stool Mini Kit, USA) was employed for a genomic DNA extraction procedure according to the manufacturer’s instruction as described formerly (7, 9, 18). The extracted DNAs were stored in -20°C till tested. nested-PCR technique was utilized to amplify a fragment of SSU 18s rRNA using 2 sets of primers as follow: (F1): 5’ - TTCTAGAGCTAATACATGCG - 3’ and (R1): 5’ - CCCATTTCCTTCGAAACAGGA - 3’ for first stage of PCR and (F2): 5’ - GGAAGGGTTGTATTTATTAGATAAAG-3’ and (R2): 5’ - CTCATAAGG TGCTGAAGGAGTA - 3’ for secondary PCR (7, 9, 19, 20). The PCR products were loaded on 1.5% agarose gel and visualized under UV light using Gel Doc device (Uvidoc, Gel Documentation System, and Cambridge, UK) (12, 13, 21, 22).
3.4. Restriction Fragment Length Polymorphism (RFLP) and Sequencing
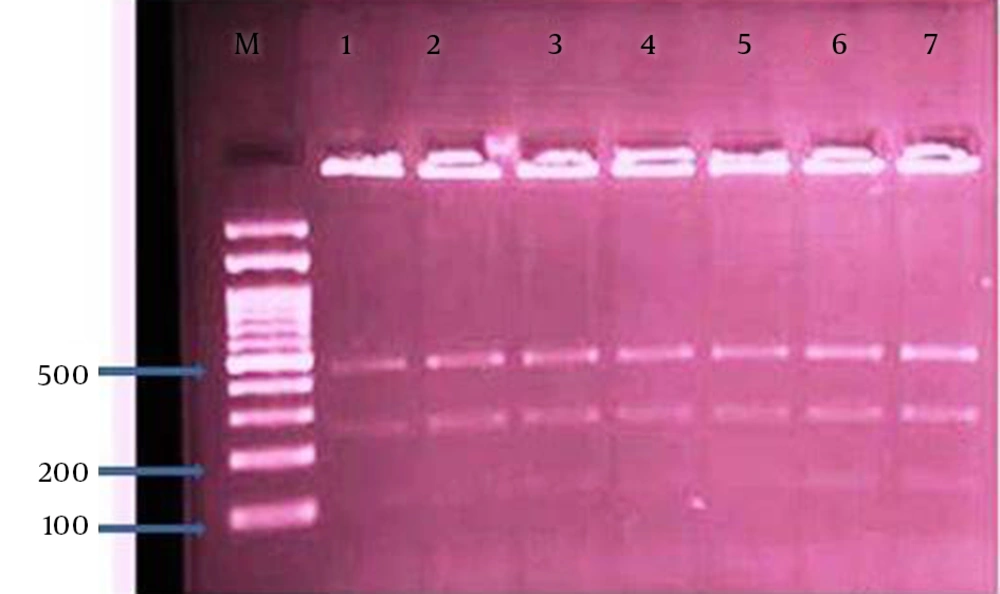
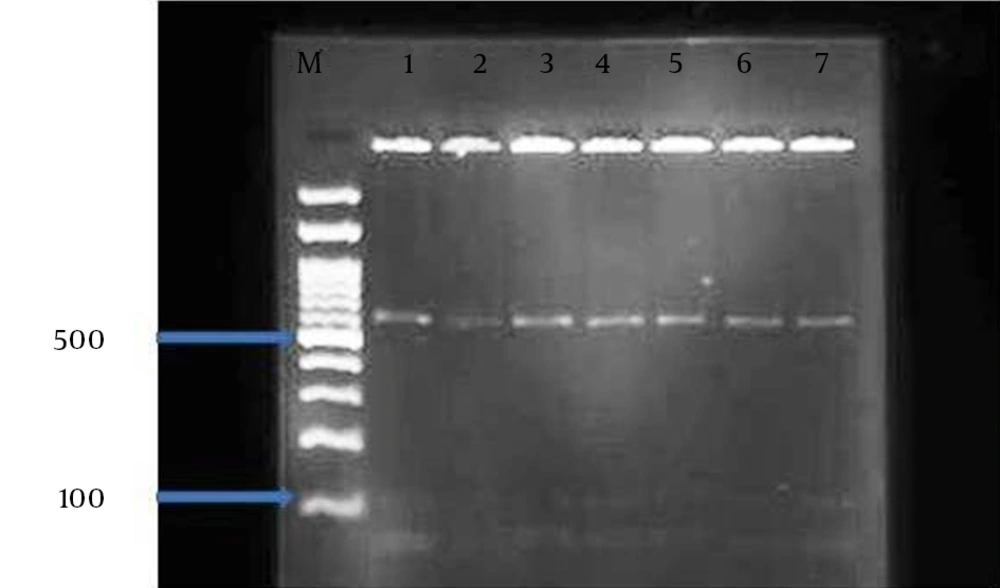
The restriction fragment length polymorphism technique was performed with digestion of secondary PCR products by SspI and VspI restriction enzymes based on the manufacturer’s guideline as explained previously (7, 9, 10, 19, 20). The restriction digestion products, after loading on 1.5% agarose gel, were electrophoresed and visualized under UV transilluminator (Uvidoc, Gel documentation system, and Cambridge, UK). The nested-PCR positive samples, after purification of using the Bioneer kit corporation (Korea), were sequenced by the same corporation. Sequence alignments were constructed by the CLUSTAL W software (http://www.ddbj.nig.ac.jp/search/clustalwe.html).
4. Results
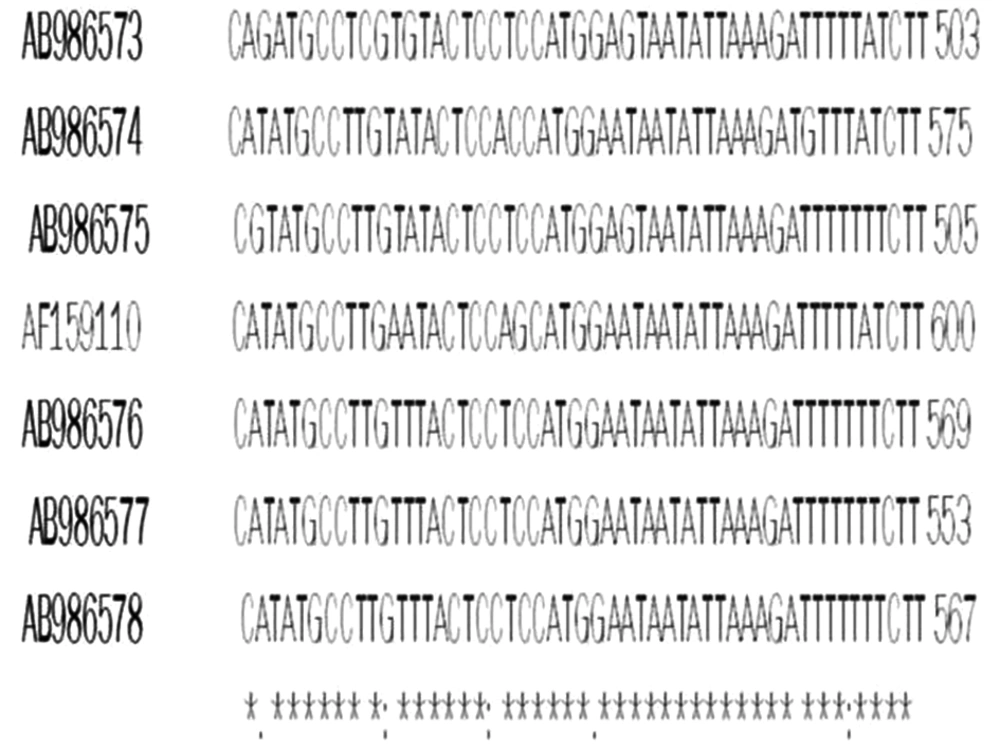
The overall prevalence of Cryptosporidium infection by modified Ziehl-Neelsen staining and nested-PCR methods were 2.1% (5/240) in the cattle. The numbers of positive cases was higher in females (4 cases) than males (1 case). PCR-RFLP analysis of nested-PCR products by employing SspI and VspI restriction enzymes revealed the presence of C. parvum in all 5 samples. It should be noted, SspI and VspI enzymes created 3 (108, 258, and 421 bp) and 2 (106 and 600 bp) visible bands, respectively (Figures 1 and 2). All these samples were sequenced by the Bioneer corporation (Korea) and C. parvum was identified. Then, the results were submitted to DDBJ/Genbank under accession numbers: AB986573-AB986577 (Figure 3).
5. Discussion
Cryptosporidium spp. belong to Apicomplexa phylum with cosmopolitan distribution (15). There are several publications on cattle cryptosporidiosis in the country of Iran (10, 11, 18, 19, 23); however, the current research is the first study regarding the prevalence and molecular characterization of Cryptosporidium spp. in the southwest region of Iran, the city of Ahvaz. In our study, the prevalence of Cryptosporidium spp. in cattle was 2.1%. Koyama et al. (2005) (24), in Japan 1.5%, and Mahami Oskouei et al. (2014) (10), in Ilam city 3.68%, (southwest of Iran) were reported to have a lower prevalence; while in China 18.82% (25), India 11.7% (26), Qazvin province 18.8% (north of Iran) (19), Urmia city 22.3% (northwest of Iran) (11), and Mashhad city 28.3% (northeast of Iran) (23), a higher prevalence was observed. The prevalence rate of Cryptosporidium spp. highly varied worldwide depending on the specific epidemiological features, locations of sampling, sample size, season, methodology, etc (8).
Recently, numerous researches on molecular characterization of Cryptosporidium spp. were successfully performed throughout the globe based on 18s rRNA PCR-RFLP technique on different samples isolated from humans, livestock, rodents, water, tissue, and so on (7, 9, 10, 18-, 25-31). The present investigation digestion of secondary PCR products using RFLP method and applying SspI and VspI enzymes, revealed C. parvum pattern in all 5 positive cases (100%). The positive cases were sequenced and submitted to DDBJ/Genbank under accession nos: AB986573-AB986577. Mahami Oskouei et al. (2014) (10), studied 217 fecal samples from cattle of western Iran and found all positive cases (3.68% - 8/217) were C. parvum. The same results were obtained by Asadpour et al. (2013) (23) in northeast of Iran, which is in accordance to our study. However, in cattle of Qazvin province, 72.6% of positive cases belonged to C. parvum and followed by C. andersoni (17.7%), C. bovis (7.8%), and new subgenotype of C. parvum (1.9%) (19). In addition, based on Mirzai et al. (2014) (11), in cattle of northwest of Iran, C. parvum and C. andersoni were identified.
In a study by Rafiei and colleagues (2014) (32), presence of parasitic agents in filtrated water, tap water, and river were examined in the western part of Ahvaz city. A total of 12 out of 44 samples (27.27%) were positive for Cryptosporidium spp. by optical microscope. Furthermore, in another survey on recreational waters of Chaharmahal va Bakhtiyari province, southwest of Iran (east of Khuzestan province), 20% of samples were positive for 18s rRNA gene of Cryptosporidium spp. (C. parvum, C. hominis and C. canis) using PCR-RFLP methods (27). A high rate of infection in water samples in each area might a result from the infection of infected animals.
6. Conclusions
In the current investigation, we utilized both microscopy and molecular methods to determine the prevalence and discernment of Cryptosporidium spp. in cattle of Ahvaz city, southwest of Iran. C. parvum was identified as the only infectious agent in the region and could be the main cause of human morbidity. More studies are required for finding the source of infection for establishing the control measures.