1. Background
Crimean-Congo hemorrhagic fever (CCHF) is an acute, viral, zoonotic disease with hemorrhagic manifestations and considerable mortality in humans. The virus is widely distributed around the world and reports of recent outbreaks have increased. Crimean-Congo hemorrhagic fever virus (CCHFV) belongs to the Nairovirus genus of the Bunyaviridae family and has a three-segmented genome: the small (S) segment encodes the nucleoprotein that encapsidates the genome and forms ribonucleoprotein (RNP) complexes (1), the medium (M) segment encodes 2 glycoproteins, and the large (L) segment encodes an RNA-dependent RNA polymerase (2). The virus is transmitted to humans either by a tick bite, by direct contact with fresh meat or blood of infected animals, or via nosocomial transmission (3-6).
Crimean-Congo hemorrhagic fever disease has been reported in some Asian, European, and African countries. Sporadic outbreaks of CCHF have been reported from Kosovo, Senegal, Turkey, Bulgaria, Iran, Pakistan, and Mauritania (2, 7-9). The predominant hosts of CCHFV include wild and domestic mammals and birds. Sheep, goats, and cattle develop high virus titers in their blood, without notable clinical signs (10). The genus Hyalomma is the main vector and reservoir of CCHFV (11, 12).
Iran is a known CCHF-endemic country, from where the disease was first reported in 1970 and the virus isolated in 1978 from ticks. Crimean-Congo hemorrhagic fever virus is detected in 27 of 31 provinces of Iran (13). Data, with respect to the gender, acquired infection in Iran have shown that CCHF infection in males is more frequent than in females, which seems to be due to male dominance in high risk professions. In this regard, farmers and their families, abattoirs, as well as healthcare workers have been considered high risk professions for acquiring CCHF (14). Although the infection rate fluctuates annually and geographically, Sistan va Baluchistan (Southeast of Iran), Khorasan (east of Iran), and Isfahan (central of Iran) provinces have the highest rates of CCHF in humans (15).
An unknown divergence time exists between 2 CCHFV species and can be estimated through the calculation of the gene evolution rate by comparing changes at the nucleotide or amino acid level of the viral genome (16). Crimean-Congo hemorrhagic fever virus has a high degree of genetic diversity, leading to 7 phylogeographic clades: clade I comprising CCHFV strains mainly isolated from West-Africa, clade II comprising CCHFV strains mainly isolated from Central Africa, clade III comprising CCHFV strains mainly isolated from South Africa and West Africa, clade IV comprising CCHFV strains mainly isolated from Middle East and Asia (this clade are divided into 2 distinct clades, Asia-1 and Asia-2), and clade V comprising CCHFV strains mainly isolated from Europe and clade VI comprising AP29 strain isolated from Greece (17, 18). In addition, the different strains of CCHFV are probably correlated with different degrees of pathogenicity (19). Consequently, understanding the molecular epidemiology of CCHFV is essential to enable the development and implementation of surveillance strategies for its control.
2. Objectives
The aim of the present study is to determine the CCHFV prevalence in ticks from Damghan (northeast of Iran), using real-time RT-RCR, and to establish a phylogenetic relationship of the tick-derived CCHFV strains circulating in Iran.
3. Methods
3.1. Study Area and Sampling
Samples were collected from Damghan county (35°30’N 54°20′E), the center of Semnan province, in Iran. All samples were collected in 2015 during the spring in 4 regions including Reza Abad, Hassan Abad, Jazan, and Damghan corresponding to 3 different climates in Damghan county (Figure 1). Ticks were subsequently collected from sheep, cow, camel and goats. In each region, animals were checked randomly for tick collection. The entire body of each animal was checked for the presence of ticks. Ticks were collected and kept alive in separate labeled vials and then transferred to the Medical entomology laboratory, school of public health, Tehran University for morphological discrimination and sex determination (20).
3.2. RNA Extraction, RT-PCR and Sequencing
All identified ticks had been kept in micro tubes and were transferred to the Arboviruses Laboratory at Pasteur institute of Iran for further analysis. Ticks were individually washed with PBS and crushed with a mortar and pestle in 200 - 300 μL of PBS. Total RNA was extracted from the samples using the RNA easy kit (QIAGEN, Viral RNA mini kit, Germany) based on the manufacturers protocol. The extracted RNAs from tick samples were screened for CCHFV RNA by RT-PCR. A master mix was prepared with QIAGEN one step RT-PCR kit (QIAGEN, Germany). In an initial screening for CCHFV RNA, a partial segment of S-gene (536-bp) were amplified using specific forward primer (5’-TGGACACCTTCACAAACTC-3’) and reverse primer (5’-GACAAATTCCCTACACCA-3’) (21, 22). Subsequently, the full length of S-gene for positive samples was amplified and completely sequenced using Sanger technique according to the previously described methods with specific primers (13, 23-25).
3.3. Phylogenetic Analysis Based on Complete S-Gene
The 4 complete S-gene sequences derived from tick samples have been deposited in GenBank (assigned accession numbers: KU707898-KU707901). The data set used for this analyses consisted of a total of 142 sequences comprised of the complete S-gene of the 4 Iranian sequences obtained in this study and 138 globally representative isolates retrieved from GenBank (http:// www.ncbi.nlm.nih.gov/genbank/). Only GenBank submissions that included both a date (between 1956 and 2015) and location of origin were considered. The alignment was performed using the MAFFT algorithm and the initial phylogenetic tree was developed by Geneious v7.1.8. Similar sequences with the same isolation year and country were omitted from the original set of 142 sequences to provide a representative dataset without duplicate sequences, which resulted in a dataset consisting of 55 sequences. A phylogenetic tree was constructed by the neighbor-joining algorithm (NJ) with HKY genetic distance model using a 1000 bootstrap replicate in Geneious v7.1.8 (26).
3.4. Nucleocapsid Protein Structure and Pairwise Identity Score
The nucloeocapsid protein (N-protein) structure of 4 CCHFV S-gene sequences obtained from ticks in Iran was predicted with approximately 65% accuracy by using EMBOSS v 6.5.7 (27). In order to tentatively classify the S-gene of CCHFV sequences obtained in this study with other CCHFV sequences, pairwise comparison and identity calculation was performed using MUSCLE, ClustalW, and MAFFT methods implemented in Sequence Demarcation Tool (SDT) v 1.2 (www.cbio.uct.ac.za/SDT) (28).
4. Results
Among 95 investigated animals, 26 (27.4%) were infested by ticks. The most infestation rate belonged to camels (60%) from the Hassan Abad region. In contrast, the lowest infestation rate belonged to sheep (17.5%) from Reza Abad (Table 1). The dispersion of different tick species collected from different districts of Damghan County located in Semnan province is shown in Table 2.
| Livestock | No. | Infestation No. | District |
|---|---|---|---|
| Cow | 8 | 3 | E |
| Goat | 8 | 3 | E |
| Cow | 12 | 2 | J |
| Camel | 10 | 6 | H |
| Sheep | 40 | 7 | R |
| Goat | 17 | 5 | R |
| Total | 95 | 26 | N A |
Detection of Infection of Tick Ectoparasites on Livestock, Studied in Damqan County, Semnan Province, Irana
| Districts/Tick Species | Rh. sanguineus | Hy. marginatum | Hy. anatolicum | Hy. asiaticum | Hy. schulzei | Hy. dromedarii | Hy. sp. | Total | Percentage (%) |
|---|---|---|---|---|---|---|---|---|---|
| Reza Abad | 43 | 1 | - | 2 | - | - | 3 | 49 | 52.69 |
| Hasan Abad | - | - | 3 | - | 2 | 20 | 25 | 26.88 | |
| Jazan | - | - | 11 | - | - | - | 11 | 11.83 | |
| Eqbalyeh | 1 | 5 | - | 1 | - | - | 1 | 8 | 8.60 |
| Total | 44 | 6 | 14 | 3 | 2 | 20 | 4 | 93 | 100 |
Dispersion of Tick Species Collected From Different Districts of Damqan County, Located in Semnan Province, Iran
4.1. Tick Species
A total of 93 ticks belonging to 6 species in 2 genera (Rhipicephalus and Hyalomma) were collected in 4 sampling areas (Reza Abad, Hassan Abad, Jazan, and Eqbalyeh), Rhipicephalus County (Table 2). The tick species were Hy. marginatum (6.45%), Hy. dromedarii (21.5%), Hy. anatolicum (15.1%), Hy. asiaticum (3.2%), Hy. schulzei (2.2%), and Rh. Sanguineus (47.3%). Meanwhile, 4.3% of ticks were female Hyalomma sp. (Table 3).
| No. | Host | Sample Location | Genus/Species | No. Collected Ticks | No. Infection to CCHFV | GenBank Accession No | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | M | Total | F | M | Total | |||||
| 1 | Sheep | R | Hy. asiaticum | 1 | 1 | 2 | - | - | - | - |
| Rh. sanguineus | 2 | - | 2 | - | - | - | - | |||
| 2 | Camel | H | Hy. dromedarii | 2 | 3 | 5 | 1 | - | 1 | KU707898 |
| Hy. schulzei | 1 | - | 1 | - | - | - | - | |||
| 3 | Camel | H | Hy. schulzei | 1 | - | 1 | - | - | - | - |
| 4 | Camel | H | Hy. dromedarii | 4 | 5 | 9 | - | - | - | - |
| 5 | Sheep | R | Rh. sanguineus | 4 | - | 4 | - | - | - | - |
| 6 | Goat | R | Hy. dromedarii | 1 | - | 1 | - | 1 | 1 | KU707900 |
| 7 | Camel | H | Hy. anatolicum | - | 3 | 3 | - | - | - | - |
| 8 | Sheep | R | Rh. sanguineus | 3 | 2 | 5 | 1 | - | 1 | KU707901 |
| 9 | Camel | H | Hy. dromedarii | 2 | 1 | 3 | - | - | - | - |
| 10 | Sheep | R | Hy. sp. | 1 | - | 1 | - | - | - | - |
| 11 | Sheep | R | Rh. sanguineus | 6 | 3 | 9 | - | - | - | - |
| 12 | Sheep | R | Rh. sanguineus | 4 | 2 | 6 | - | - | - | - |
| 13 | Goat | R | Rh. sanguineus | - | 2 | 2 | - | - | - | - |
| 14 | Sheep | R | Hy. marginatum | - | 1 | 1 | - | - | - | - |
| Rh. sanguineus | 5 | 1 | 6 | - | - | - | - | |||
| 15 | Goat | R | Rh. sanguineus | 1 | 3 | 4 | - | - | - | - |
| 16 | Cow | J | Hy. anatolicum | 3 | 3 | 6 | 1 | - | 1 | KU707899 |
| 17 | Goat | R | Hy. sp. | 1 | - | 1 | - | - | - | - |
| Rh. sanguineus | 3 | - | 3 | - | - | - | - | |||
| 18 | Goat | R | Rh. sanguineus | 2 | - | 2 | - | - | - | - |
| 19 | Cow | J | Hy. anatolicum | 2 | 3 | 5 | - | - | - | - |
| 20 | Cow | E | Hy. marginatum | - | 1 | 1 | - | - | - | - |
| 21 | Cow | E | Rh. sanguineus | - | 1 | 1 | - | - | - | - |
| 22 | Camel | H | Hy. dromedarii | 2 | 1 | 3 | - | - | - | - |
| 23 | Cow | E | Hy. marginatum | - | 2 | 2 | - | - | - | - |
| Hy. asiaticum | - | 1 | 1 | - | - | - | - | |||
| 24 | Goat | E | Hy. sp. | 1 | - | 1 | - | - | - | - |
| 25 | Goat | E | Hy marginatum | - | 1 | 1 | - | - | - | - |
| 26 | Goat | E | Hy. marginatum | - | 1 | 1 | - | - | - | - |
| Total | 26 livestock | 4 districts | 6 species | 52 | 41 | 93 | 3 | 1 | 4 | - |
Details of Collected Ticks and Their Infection to CCHFV Studied in Damqan County, Semnan Province, Irana
4.2. Crimean-Congo Hemorrhagic Fever Virus Prevalence in the Tick Populations
The CCHFV RNA was identified in 4 tick samples (4.3%). The infected samples belonged to a female Rh. sanguineus, a female Hy. dromedarii, a female Hy. Anatolicum, and a male Hy. dromedarii collected from sheep, camel, cow, and goat, respectively. A total of 3 positive tick samples were female ticks (75%) and 1 male tick (25%). It is notable that 52 of 93 tick samples (55.9%) were female. The infection rate of female and male ticks is proportional with the female and male ectoparasite samples that were collected (Table 3).
4.3. Phylogenetic Analysis of Full-Length S-gene Crimean-Congo hemorrhagic Fever Virus Strains
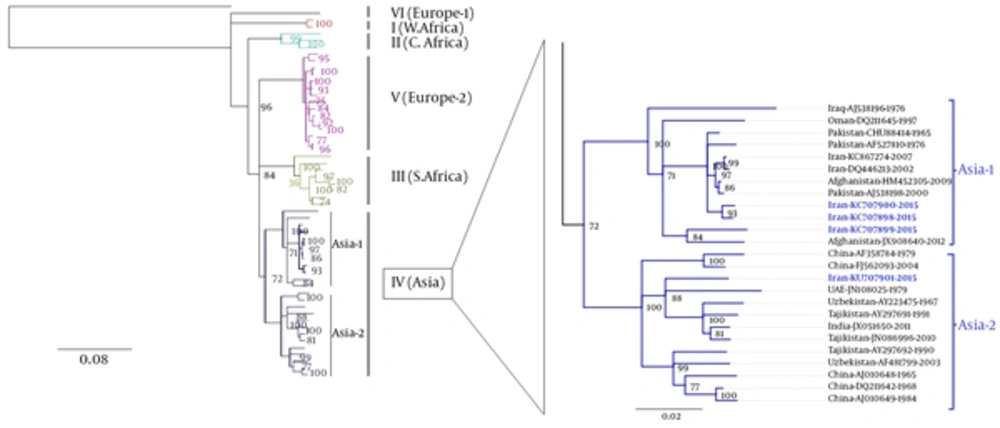
The phylogenetic tree was deduced from 51 previously published CCHFV strains and 4 CCHFV sequences in this study. The phylogenetic tree inferred from the complete coding region (ORF) of CCHFV S-gene sequences revealed 7 distinct clades (Figure 2). All 7 main clades were confirmed by the maximum likelihood inference and supported by bootstrap values. The phylogenetic analyses indicated that all 4 recent CCHFV sequences obtained from tick samples fell into clade IV. Meanwhile, CCHFV sequences obtained from 2 Hy. dromedarii (KU707898 and KU707900) and 1Hy. anatolicum (KU707899) grouped within clade IV (Asia-1), whereas the remaining CCHFV sequence obtained from Rh. sanguineus (KU707901) clustered with other CCHFV strains within clade IV (Asia-2).
4.4. Structural Comparison of Iranian Crimean-Congo Hemorrhagic Fever Virus N-Proteins and Identity Matrix
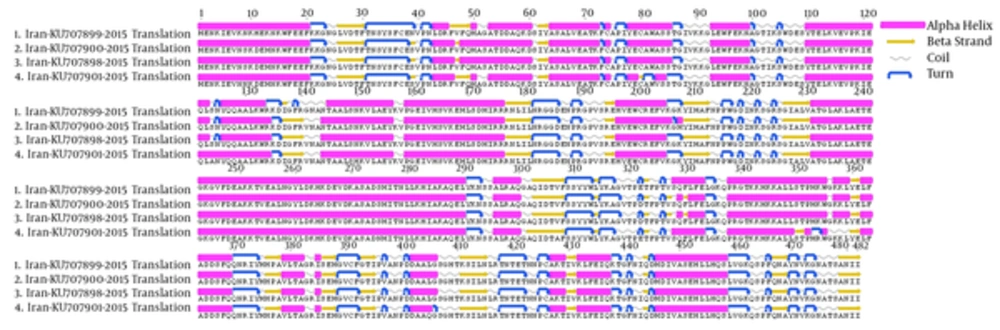
The S-gene of CCHFV encodes 482 amino acids (102 alpha helices, 89 beta strands, 146 coils, and 147 turns), forming the monomeric N protein, which when combined with CCHFV RNA forms the viral nucleocapsid structure (29). The overall protein structural topology from 4 CCHFV strains obtained in this study was similar. Interestingly, comparison of the primary sequences of 4 CCHFV strains indicated 16 a.a. differences among 482 positions, however, only 8 a.a. changes caused secondary structure alternation. The main secondary structural difference was the 5 divisions of long alpha helixes in strains KU707899 and KU707901 at a.a. positions 48 to 60, strain KU707901 at a.a. positions 78 to 85, strains KU707899, KU707900 and KU707898 at a.a. positions 110 to 134, strains KU707900 and KU707898 at a.a. positions 328 to 333, and strain KU707901 at a.a. positions 338 to 355, into 2 shorter alpha helices. One beta sheet was shortened and a turn in secondary structure occurred at a.a. positions 137 to 141 in strain KU707899 and an extended alpha helix at a.a. position 208 after turn in strain KU707900. An alpha helix changes to coil at a.a. positions 357 to 359 in strain KU707901. One alpha helix was shortened and a turn in secondary structure occurred at a.a. position 406 in strain KU707901 (Figure 3).
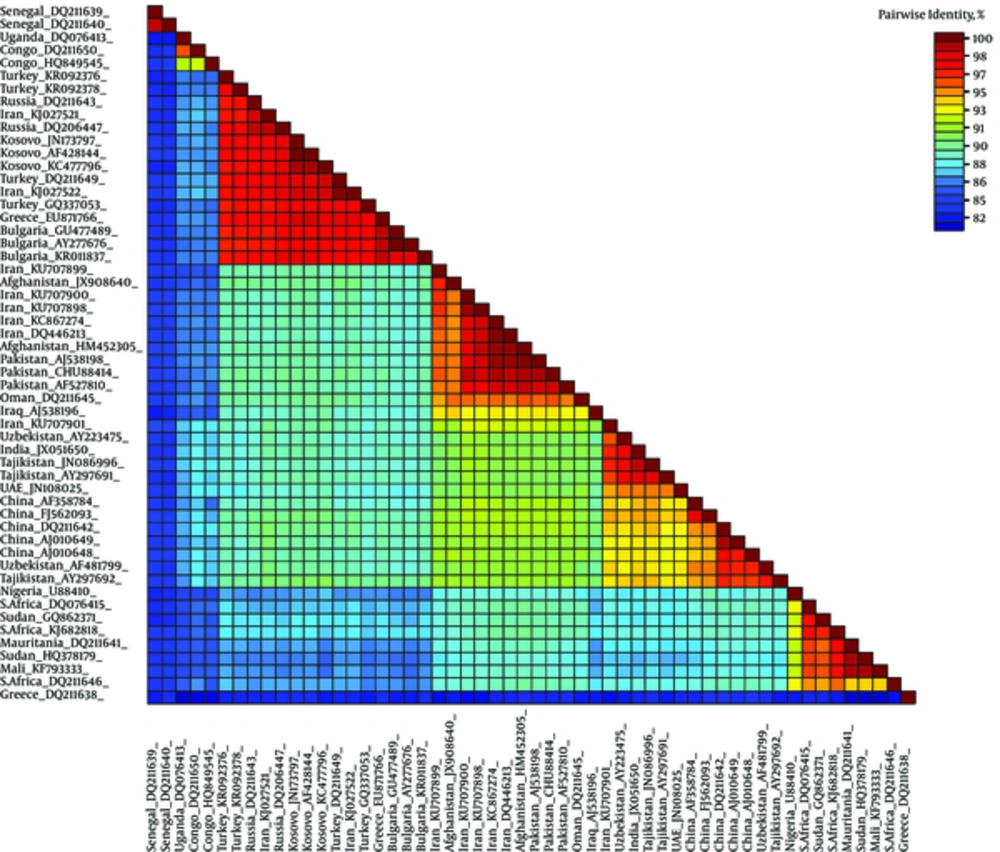
The SDT analysis showed pairwise identity scores using a color-coded matrix and provided an insight into the overall relationship between sequences in the dataset. All 3 alignments resulted in similar demarcation patterns (data not shown). Only the MAFFT alignment identity matrix is shown. Obvious demarcation between different clades of CCHFV is distinguishable (Figure 4).
Each colored cell represents the percentage identity score between 2 sequences (one indicated horizontally to the left and the other vertically at the bottom). A color key indicates the correspondence between pairwise identities and the colors displayed in the matrix. High genetic identity (100%) is indicated (dark red color). In contrast, genetically distant sequences are also coded (blue color)
5. Discussion
Several arboviruses are circulating in Iran (30-35), however, CCHF human causes are considered as the most severe arbovirus in Iran with considerable mortality rate each year (36, 37). The seroprevalence of CCHFV in humans and animals in Iran was investigated for several years (38). Approximately, 100 human CCHF cases per year have been reported in Iran since 2000 (39). Given the essential role of ticks as the vectors and reservoirs of human and animal diseases, they are the subjects of many studies in Iran (40). Our results showed that 6 tick species are infesting livestock in the investigated areas. In addition, we demonstrated the presence of CCHFV in 2 Hy. dromedarii, 1 Rh. sanguineus, and 1 Hy. anatolicum from Damghan county in Semnan province. The detection of CCHFV in ticks is important, because different tick species are responsible for transmission of CCHFV and are available in Iran, which might increase the risk of CCHFV transmission to humans (41).
In a recent study, tick species infesting livestock in a neighboring province of Semnan province, Mazandaran province, was screened for CCHFV by RT-PCT. As a result, CCHFV RNA was detected in Rh. sanguineus, Rh. bursa, Ixodes ricinus, Boophilus annulatus, Haemaphysalis punctate, and H. numidiana (42). In our report, Rh. sanguineus was also identified as a positive tick reservoir for CCHFV. Both the phylogenetic analysis and identity matrix analysis, based on the full length of S-gene, demonstrated the clustering of CCHFV strains within 7 distinct clades, and revealed the close genetic relationship of 4 CCHFV strains obtained in the current study with other CCHFV strains in clade IV, including Asia-1 and Aisa-2.
The variations of CCHFV genome sequences combined with sample collection dates and locations can be used to identify the source and the evolutionary history of circulating strains. The results of our genome-based epidemiology revealed that 2 CCHFV variants are circulating in this part of Iran, which is in accordance with previous phylogenetic data (30). Grouping of the complete CCHFV S-segment sequences obtained in the current study are in conformity with the previous reports (36, 37).
Understanding of N protein is noteworthy, as the N-protein interacts with the cellular antiviral defense factor MxA and was recently shown to have a role as a substrate for the apoptosis mediator caspase-3 (43, 44). Thus, amino acid substitutions may lead to protein structure modification that would consequently alter its ability to combat cellular defense mechanisms against CCHFV and also influence its functions such as RNA binding and oligomerization, which are required for virus replication. Additional studies are required to further understand how the host antiviral defense against circulating CCHFV strains in Iran is activated, which will enable therapeutic molecules to be designed and used for human use (45). In addition, the identification of pathogen transmission mechanisms in ticks may provide the opportunity to disrupt transmission routes and may contribute to the development of vaccines against tick-borne diseases (46) including strains of CCHFV that circulate in Iran.
6. Conclusion
To conclude, the continuous emergence of CCHFV is a public health concern in Iran (47). Crimean-Congo hemorrhagic fever virus is circulating in several countries surrounding Iran e.g Turkey (48), Pakistan (49), and Afghanistan (50) with sporadic outbreaks, which demands collaboration between involved countries to cope with CCHFV re-emergence. Accordingly, public health authorities in the mentioned countries should pay more attention to vector control measures to reduce the density of CCHFV vectors.