1. Background
Lactic Acid Bacteria (LAB) have been classified as GRAS (generally recognized as safe), as they are part of the human commensal microflora and generally present in many fermented and nonfermented food products. This group of bacteria is of significant importance in the dairy and biotechnology industries, where they are used as starter cultures for dairy fermented food products, human, and animal health products as well as animals feed inoculants. Besides the nutritional values, the beneficial effects of probiotics on human and animal health, reduction of pathogens in the food chain, and growth performance have increasingly been highlighted (1, 2).
Generally, probiotic bacteria should possess the essential properties such as resistance to stressful conditions of stomach and upper intestine where acid and bile are the most detrimental factors effecting growth and viability (3, 4). Apart from these characteristics, the antagonistic effects and adherence to the intestinal epithelium are the characters considered essential of an effective probiotic microorganisms (3, 5). Moreover, prior to the application of a LAB as a probiotic source in the food and feed industry, one of the prerequisites for ensuring consumer safety (5, 6) is its assessment of its safety properties, including lack of antibiotic resistance and in vivo safety. These evaluations could lead to improvements in quality and functional properties of the probiotic products.
Dairy products are well known for their dietary health benefits and are considered a rich source of beneficial bacteria, including Lactic Acid bacteria. Traditionally made dairy products like yoghurt, butter, and sour butter milk prepared from ewe milk in a rural area in East Azerbaijan of Iran has been quite well known and popular among its local. In the last couple of years, these traditionally made dairy products gained a lot of importance throughout the country not only due to their flavor, texture, and a greater shelf life, but also due to their well-known health benefits.
2. Objectives
This study aimed to investigate in detail the probiotic potential of LAB flora present in ewe milk. To our knowledge, this is the first report in Iran on the isolation and characterization of beneficial bacteria in traditional dairy products made from ewe's milk.
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
Two Lactobacillus strains isolated from ewes milk in a previous study (7) were used in the study. The isolates were grown in MRS broth (HiMedia, India) at 37˚C for 24 - 48 hours. Other Gram negative and Gram positive pathogens used in study were grown in brain-heart infusion broth (BHI, Oxoid, UK) except for Haemophilus influenzae b, which was grown in SBHI (supplemented BHI). The strains were maintained at 4˚C and renewed every week for short-term preservation, while for long-term conservation cell suspension of the isolates in 10% skim milk with 20% sterile glycerol were stored at -70˚C.
3.2. Phenotypic Characterization of Selected Probiotic Strains
Selected isolates were tested for their motility, spore forming ability, oxidase test, acid and gas production from glucose, and growth at 15˚C and 45˚C. Arginine hydrolysis test was performed using MRS broth without glucose and meat extract containing 0.3% arginine and 0.2% sodium citrate instead of ammonium citrate. Arginine MRS medium and Nessler’s reagent were used in order to see ammonia production from arginine (8).
3.3. Genus and Species Identifications by PCR Reactions
Both the isolates were identified to genus level by PCR (polymerase chain reaction) using 16S rRNA gene sequencing. PCR parameters and universal primers 7f (5'-AGA GTT TGA TYM TGG CTC AG-3') and 1510r (5'-ACG GYT ACC TTG TTA CGA CTT-3') were used as described by Satokari and colleagues (9). The amplified products were sequenced and the obtained sequences were aligned to 16S rRNA gene sequences in the GenBank data base using the BLAST algorithm. The selected isolates were identified to species level by the use of specie specific primers. The primer pairs used were brevisF 5′-CTTGCACTGATTTTAACA-3′, brevisR 5′- GGGCGGTGTGTACAAGGC-3′ for L. brevis and pentF 5′-CAG TGG CGC GGT TGA TAT C-3′ and pentR 5′-TCG GGA TTA CCA AAC ATC AC-3′ for L. pentosus (10). All reactions included positive (L. brevis PTCC and L. pentosus ATCC 8407) and negative (Enterococcus faecalis PTCC 1394) controls.
3.4. Bile Tolerance
The bile tolerance of the respective isolates was examined by slightly modifying the method of Walker and Gilliland (11). MRS broth medium supplemented with 0.3%, 0.5%, 1%, 2%, and 3% Oxgall (Sigma, UK) were inoculated with active cultures (1% v/v) and incubated at 37˚C. Control consisted of MRS broth with the respective bile salt concentrations. Growth was monitored at 0, 4, and 8 hours by recording absorbance at 650 nm. All tests were run in duplicate and mean results were recorded. Formula of Gopal et al. calculated coefficient of inhibition (Cinh) (12).
Formula 1.
Cinh = ∆T8-T0 Control – ∆T8 - T0 Treatment / ∆T8 - T0 Control
Where, ∆T8 - T0 represents the difference in absorbance at time zero (T0) and after 8 hours (T8). Cinh of less than 0.4 was considered significant for the isolates to be considered a suitable probiotic candidate.
3.5. Bile Salt Hydrolysis
Hydrolysis of the bile by the selected probiotic isolates was determined by the procedure described earlier (13). Briefly, the sterile filter disks impregnated in overnight grown cultures of the selected strains were placed on MRS agar plates supplemented with 0.5% taurodeoxycholic acid sodium salt (Sigma, UK). The plates incubated at 37˚C for 48 hours and observed for precipitation zones around the disks.
3.6. Survival in Simulated Gastric Content
The resistance of the selected lactobacillus isolates to simulated gastric content was evaluated by slight modifications in Beumer et al. method (14). Simulated gastric juice medium (pH 3.0) was prepared by adding pepsin (13.3 mg/ L), lysozyme (0.1 mg/ L), porcine bile (0.05 mg/L), and 0.5 % sodium chloride. All enzymes used in study were purchased from Sigma, UK. The washed cells of the probiotic strains were added (1% v/v) to the gastric juice medium at 37˚C with constant stirring. The survival of tested bacteria was determined after 0, 10, 20 and 30 min by pour plate method and the number of viable organisms (cfu/mL) recorded. The bacterial survival was calculated as follows:
Formula 2.
R = of cells at 10 min/average of cells at 0 min
According to the formula, R = 1 when no effect on the growth and survival of bacteria is seen, while a ratio of 0.5 indicated a loss of 50% viability. Ratios greater than 1 indicate bacterial growth.
3.7. Survival in Simulated Upper Intestine Content
Viability of the selected lactobacillus isolates in simulated upper intestine content (UIJ) was evaluated by the Charteris et al. method (15). UIJ was prepared by adding 1 g/L of bile salt in 1 M PBS solution (pH 8.0) containing pancreatic enzyme (Pancrex, UK) at final concentrations of 1 g/L. Washed cell suspension of the tested strains was added at concentrations of 1% v/v to the above mentioned medium and incubated at 37˚C. The survival of the tested bacteria (cfu/mL) at 0, 4, 8 and 24 hours was determined using the above mentioned formula.
3.8. Antimicrobial Spectrum
The antimicrobial effects of selected LAB against Gram positive and negative pathogens were examined by agar-well diffusion method described earlier (16). The antimicrobial activity was recorded as appearance of clear zone around the wells and their zone diameter (mm) was recorded.
3.9. Cholesterol Removal
Cholesterol reducing ability of the selected isolates was determined by adding filter sterilized water-soluble cholesterol (100 μg/mL) to MRS broth containing oxgall 0.3 % and taurocholic acid (Sigma-Aldrich, UK), respectively. The tube were inoculated with the selected strains (1% v/v) and incubated at 37˚C. Control sample included non-supplemented MRS broth. After 0, 2, 4 and 8 hours samples were taken and the residual cholesterol of supernatants determined using a modified colorimetric method described by Rudel and Morris (17). The cholesterol lowering ability was determined by measuring the absorbance at 550 nm, and calculating percentage from the following Equation:

Where A = % cholesterol removed, B = absorbance of blank, and C = absorbance of cell supernatant.
3.10. Autoaggregation and Coaggregation Assay
The extent of autoaggregation by the selected probiotic isolates was assessed by slight modifications of the method described by Reniero and his colleagues (18). Overnight grown cultures of the tested isolates were harvested by centrifugation (5000 x g, 20 minutes, 4˚C). The pellet was washed thrice with sterile distilled water and suspended in PBS (pH 7.0) to obtain an OD (600 nm) of 0.6. The tubes were incubated at 20˚C and the absorbance at 600 nm of the cellular suspensions was monitored every 1 hour for a period of 4 hours. The percentage of autoaggregation was calculated by the following Formula:
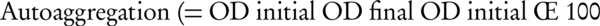
Where, OD initial = OD at time 0, and OD final = OD at each time after beginning this assay (t = 1, 2, 3 or 4 hours).
Coaggregation assay was performed by mixing equal volumes of washed cell suspension of selected probiotic isolates with equal volume of overnight grown cultures of L. monocytogenes (RTCC 1298), and S. pneumoniaea (local isolate), respectively. The tubes were incubated at room temperature and absorbance at 600 nm was measured at 4 hours. Appropriate controls included pure cultures of bacterial cells suspension in PBS, individually. The percentage of coaggregation was calculated following the Equation of Handley et al. (19):
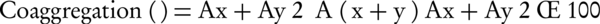
Where, Ax and Ay represent absorbance (A) of each of the 2 strains in the control tubes, and (x + y) the mixture.
3.11. Adhesion Assay
The adherence of selected probiotic isolates to Caco-2 cell lines was determined by the method followed by Pan and his colleagues (20). In brief, the Caco-2 cells (ATCC 7348406) seeded on glass cover slips were inoculated with 120 μL of respective overnight bacterial cultures (approximately108 cells/ mL). After incubation at 37˚C for 1 hour, the treated cells were washed thrice with PBS (pH 7.0) and fixed with methanol for 5 minutes at room temperature. The cells were stained with May–Grunwald–Giemsa (Sigma, UK) for 30 minutes, washed thoroughly until all color was drained off, dried overnight, and analyzed under oil immersion lens (100×). The number of cells adhered were obtained by scoring adhesion to 100 random eukaryotic cells and recording results as nonadhesive, adhesive, and strongly adhesive, when < 5 bacteria/100 cells, 6 – 40 bacteria/100 cells and > 40 bacteria/100 cells were adhered, respectively (21).
4. Results
In a previous study, we had isolated a number of lactic acid bacteria from ewe milk in a rural area of Iran. In this research study, we selected two of the isolates identified as L. brevis LB32 and L. pentosus LP05 by their carbohydrate fermentation profiles for detailed investigations. The 2 isolates were catalase and oxidase negative, heterofermentative (produced acid and gas from glucose), nonmotile and grew well at 15˚C, but were unable to grow at 45˚C. Two isolates were confirmed to species level by 16S rRNA gene sequencing using universal primers. The Blast search showed 99% and 97% sequence similarity with L. brevis (NCBI GenBank accession no KF975703 and L. pentosus (NCBI GenBank submission ID1685019), respectively. The isolates subjected to species specific primers showed a DNA product of 1340 in L. brevis LB32, while L. pentosus LP05 indicated a DNA band of approximately 218 bp.
Table 1 depicts the survival of the tested strains in the presence of variable concentrations of bile salt at different time intervals. Both isolates were considered bile tolerant as their coefficient of inhibition (Cinh) appeared less than 0.4% up to 1% of bile salt. According to the results, Cinh of L. brevis LB32 was higher during the initial 4 hours while it decreased later and significant reduction was observed at 8 hours. The contrasting results were recorded for L. pentosus LP05, which showed lower Cinh values during 4 hours compared to the greater values at 8 hours. The bile tolerance of the isolates is indicative of their possible survival in small intestine.
| Bile salt, % | Time, h | Coefficient of inhibition (Cinh) | |
|---|---|---|---|
| L. brevis LB32 | L. pentosus LP05 | ||
| 0.3 | 4 | 0.05 | 0.02 |
| 8 | 0.03 | 0.04 | |
| 0.5 | 4 | 0.08 | 0.08 |
| 8 | 0.11 | 0.15 | |
| 1 | 4 | 0.21 | 0.22 |
| 8 | 0.17 | 0.36 | |
| 2 | 4 | 0.57 | 0.61 |
| 8 | 0.55 | 0.70 | |
| 3 | 4 | 0.78 | 0.0 |
| 8 | 0.57 | 0.0 | |
| 0.3 | 4 | 0.05 | 0.02 |
| 8 | 0.03 | 0.04 | |
Coefficient of Inhibition in Medium With Variable Concentrations of Oxgall After Different Time Period
A significant reduction in the viability of the tested Lactobacillus strains was recorded in simulated gastric juice contents with elapse of time. The survival percentage within 30 minutes under these conditions was still higher than 50% (Table 2). The effect of simulated intestine contents on the selected lactobacilli isolates indicated tolerance of the tested isolates to alkaline pH values (pH 8.0). The survival rate of both isolates was significantly higher in simulated upper intestine conditions compared to gastric conditions where acidic pH was prevalent. Although none of the isolates was able to resist the alkaline conditions immediately after addition and their viability was extremely low at time zero; however, the growth accelerated significantly within 4 hours reaching its maximum within 24 hours of incubation. Comparatively, L. brevis LB32 appeared more resistant than L. pentosus LP05, as it showed higher viability in both the simulated gastric and upper intestine contents.
| Probiotic Isolates | Survival (R) | |||||||
|---|---|---|---|---|---|---|---|---|
| Simulated Gastric Content | Simulated Upper Intestine | |||||||
| 0 | 10 | 20 | 30 | 0 | 4 | 8 | 24 | |
| L.brevis LB32 | 1.0 | 0.9 | 0.81 | 0.70 | 0.53 | 1.51 | 1.69 | 2.0 |
| L.pentosus LP05 | 1.0 | 0.8 | 0.72 | 0.67 | 0.61 | 1.20 | 1.36 | 1.6 |
The Viability of Selected Probiotic Isolates in Stimulated Gastric and Upper Intestine Content at Different Time Intervals
Table 3 shows the results of antagonistic activity of selected probiotic isolates against different Gram positive and negative pathogens. The isolates demonstrated different level of inhibitory action against the pathogens as obvious by their zone diameters. L. brevis LB32 showed wider spectrum of inhibition compared to L. pentosus LP05, as it also inhibited the growth of S. dysenteriaea and K. pneumoniae. Both isolates were unable to inhibit the growth of H. influenzae b and P. aeroginosa, while S. pneumoniae appeared to be the most sensitive strain as apparent by the highest zone of inhibition against this pathogen.
| Indicator strains | L. brevis LB32 | L. pentosus LP05 |
|---|---|---|
| Haemophilusinfluenzae b (ATCC 10211) | 0 | 0 |
| Klebsiellapneumoniae (PTCC 1053) | 16 | 0 |
| Listeria monocytogenes (RTCC 1298) | 21 | 24 |
| Pseudomonas aeruginosa (local isolate) | 0 | 0 |
| Salmonella enteritidis (local isolate) | 17 | 21 |
| Salmonella typhi (local isolate) | 14 | 16 |
| Shigelladysenteriaea (local isolate) | 19 | 0 |
| Shigelladysenteriaea (local isolate) | 16 | 0 |
| Staphylococcus aureus (RTCC 1112) | 12 | 13 |
| Streptococcus pneumoniae (local isolate) | 27 | 25 |
The Zone of Inhibition (mm) Demonstrated by Selected Probiotic Isolates against Gram Positive and Negative Pathogens
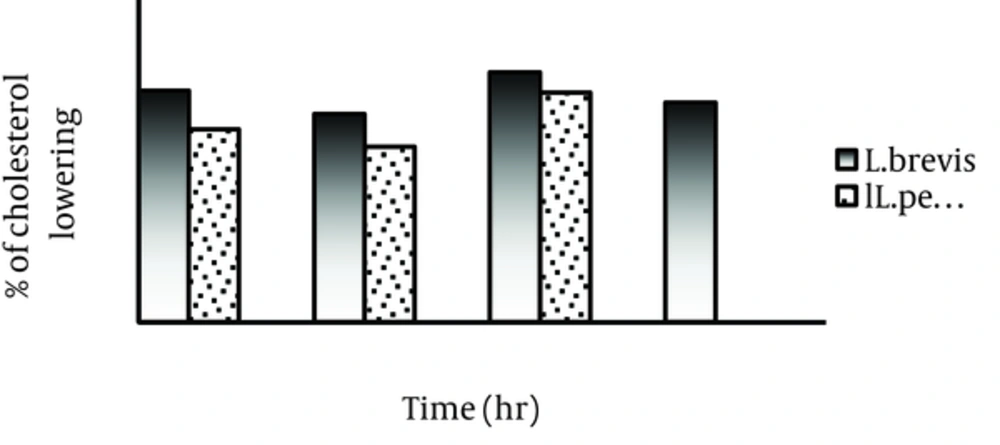
The percentage of cholesterol assimilation during different time periods revealed wide variations in the mentioned property of the tested isolates and variable levels of cholesterol reduction were observed (Figure 1). L. brevis LB32 assimilated highest level of cholesterol in the presence of oxgall 0.3% during 8 hours, while L. pentosus LP05 exhibited lower cholesterol reducing ability in the same condition. Cholesterol assimilation by both the tested isolate was higher in the presence of oxgall (0.3%) compared to the presence of same concentrations of taurocholic acid.
The autoaggregation and coaggregation properties of the selected lactobacillus strains were determined (Table 4). According to the results, L. pentosus LP05 demonstrated strong autoaggregation phenotype and approximately 70% of its cells were able to aggregate within 4 hours of incubation. Although L. brevis LB32 also showed autoaggregation ability, its percentage of aggregation was lower than that of L. pentosus LP05. Moreover, L. brevis LB32 was not able to show any aggregation during the initial first hour of incubation, however after 2 hours of incubation an observable aggregation property was observed.
| Probiotic Isolates | Cholesterol Reduction, %% | Auto-Aggregation, % | Co-Aggregation, | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.3% Oxgall | 0.3% TCA | 1h | 2h | 3h | 4h | L.M | S.P | |||||
| 2h | 4h | 8h | 2h | 4h | 8h | |||||||
| L. brevis LB32 | 79.6 | 71.5 | 81.2 | 54.3 | 65.7 | 65.8 | -ve | 21.9 | 38.9 | 56.8 | 51.24 | 37.46 |
| L. pentosus LP05 | 66.3 | 66.6 | 69.0 | 61.4 | 66.3 | 62.4 | 12.6 | 44.6 | 64.7 | 70.2 | 48.11 | 54.06 |
Percentage of Cholesterol Reduction, Aggregation and Coaggregation by Selected Probiotic Isolates at Different Time Interval a
Coaggregation ability of the two isolates with the selected pathogens like L. monocytogenes and S. pneumoniae was tested and results recorded in percentage of coaggregation. According to the results, coaggregation ability is directly related to autoaggregation phenotype, as higher co-aggregation percentage was seen in strains possessing higher aggregation ability. L. pentosus LP05 has a higher autoaggregation and coaggregation ability, compared to L. brevis LB32. Comparing the two strains ability to coaggregate, it appeared that L. brevis LB32 could coaggregate more with L. monocytogenes than with S. pneumoniae, while L. pentosus LP05 showed enhanced coaggregation with S. pneumoniae compared to the other pathogens. Both probiotic isolates in this study were able to adhere to Caco-2 cells. According to the results, the numbers of adhesive bacteria of L. brevis LB32 and L. pentosus LP05 to Caco-2 cells were approximately 64 and 71, respectively. The results of this assay indicated a direct correlation of adhesion to aggregation phenotype.
5. Discussion
Dairy products are well known for their dietary health benefits and are considered a rich source of beneficial bacteria such as lactic acid bacteria. Traditionally made dairy products like cheese, yoghurt, butter, and sour butter milk prepared from ewe milk in rural areas in East Azerbaijan of Iran is quite well known and popular among their locals and people from big cities in the country. These traditionally made dairy products have gained a lot of importance because of their flavor, texture, greater shelf life and mainly their well-known health benefits. The present investigation highlights the important characteristics of two Lactobacillus strains isolated from traditionally made sour butter milk made from ewe milk. To our knowledge, this is the first report on the probiotic characterization of beneficial bacteria in ewe milk made traditional dairy products in Iran.
In this study, the efficacy of two Lactobacillus strains namely L. brevis LB32 and L. pentosus LP05 was evaluated and scrutinized in order to highlight their beneficial health effects. According to numerous reports, an essential criterion for selection of probiotic strains is their acid and bile resistance (22, 23). The acid resistance of the selected isolates in study was in accordance with the reports of Garriga et al. (24) who suggested that resistance to pH value of 3.0 for 2 hours is an optimal value for selecting probiotic bacteria. None of the isolates resisted pH values of lower than 2.0, which could be explained by the fact that probiotic bacteria are not exposed to such extreme pH values in the stomach as are buffered by food or other carrier matrix molecules after consumption (25).
Both the isolates in study were bile resistant, which is indicative of their possible survival in small intestine. Moreover, resistance to bile salt of the isolates could be attributed to their ability to produce bile salt hydrolase (BSH). BSH is an enzyme responsible for deconjugation of bile acids, which are less soluble and less efficiently reabsorbed from the intestinal lumen than their conjugated counterpart. Therefore, deconjugation of bile salts could lead to decrease in serum cholesterol (26). There are different possible mechanisms underlying the ability of LAB to remove cholesterol from media, including assimilation (27), enzymatic hydrolysis of conjugated bile salts, and coprecipitation of cholesterol with deconjugated bile salt (28, 29). As reported, a significant relationship exists between cholesterol assimilation by lactobacilli and their degree of bile deconjugation (27). In agreement to these reports, the isolates in our study showing the ability to hydrolyze bile salt also showed significant level of cholesterol reduction.
The ability of the isolates in study to survive stimulated gastric and intestine content is in agreement with other previously reported probiotic bacteria (30, 31). The greater resistance of L. brevis LB32 in simulated upper intestine contents is in contrast to the reports of Charteris et al. (15), who reported greater sensitivity of Lactobacillus spp to simulated intestine content. Another important characteristic of the 2 lactobacillus isolates in study was their antibacterial activity against a number of Gram negative and Gram positive pathogens. Such inhibitory activities have been reported earlier in a numbers of LAB such as L. brevis and L. acidophilusL. sake, L. pentosus, etc.(32, 33). The antibacterial potential of these bacteria has been attributed to its ability to produce H2O2, acids, phages, bacteriocins, and bacteriocin like substances (34).
The aggregation property of some LAB is considered to play an important role in colonization of the probiotic strains in gastro intestinal tract, and has also appeared necessary for adhesion to intestinal epithelial cells (35, 36). Similarly the coaggregation abilities may form a barrier that prevents colonization by pathogenic bacteria. L. brevis LB32 and L. pentosus LP05 were able to demonstrate aggregation and coaggregation phenomenon at different time intervals. The ability to adhere to epithelial cells and mucosal surfaces has been suggested to be an important property of many bacterial strains used as probiotics. In most cases, aggregation property is related to cell adherence (21). This phenomenon was also observed during our studies, as the bacteria demonstrating higher aggregation and coaggregation properties showed higher adherence properties to Caco-2 cell lines.
To conclude, the traditionally made dairy products from ewe milk, especially in a rural area of Iran are rich sources of probiotic lactobacilli. As our study indicates that L. brevis LB32 and L. pentosus LP05 isolated from the mentioned dairy product possess significant probiotic attributes, which could be exploited for production in future. However, further studies on the safety assessment, including the presence of antibiotic resistance genes and in vivo testing on an appropriate animal model is essential before recommending them for use in human or animal.