1. Background
Acute respiratory infections (ARIs) are one of the important causes of morbidity and mortality worldwide. World Health Organization has estimated that annually, ARIs are responsible for the death of four million people worldwide (1). The highest percentage of these deaths happens in children in developing countries (2). Symptoms of upper respiratory infections (URI) most commonly include rhinorrhea, sore throat, nasal congestion, sneezing, cough, headache, fever, and in some cases malaise and myalgia; in lower respiratory infections (LRI), the symptoms are pneumonia, bronchitis and bronchiolitis. These symptoms have been seen either in viral or bacterial infections, so it has become a challenge for physicians to differentiate viral causes from bacterial ones, which may decrease the excessive uses of antibiotics (3-5).
Distinguishing the etiology also allows the physician to tell the patients about the assessment and the way of prevention of transmission to others (4). Investigations on respiratory infections have been started since 1933 with isolation of influenza viruses and after three decades other various respiratory viruses have been discovered, but some have remained undetermined because of the low sensitivity of methods (6). Usually, traditional viral culture, generally in combination with serology and direct immunofluorescent (DIF) is the key method for laboratory diagnosis of viral respiratory infections; however, these methods have lower sensitivity, need specific technical expertise and advanced labor instruments, are expensive and are not useful for on-time diagnosis (7).
In some particular situations, the emergence of respiratory viruses has an important impact on health systems worldwide, as happened with the emergence of severe acute respiratory syndrome (SARS)-coronavirus in 2003 which needed emergency diagnosis (8). The development of molecular methods for direct detection of viruses has recently been progressed (9). Polymerase chain reaction (PCR) has become an accepted tool in research since 1984 by Kary Mullis et al. (10) and is being introduced into laboratories (11). Singleplex PCR assays amplify one target in different reactions; they are resource intensive and expensive. Multiplex PCR uses a mixture of several distinctive primer pairs in one amplification reaction at the same time (12). Nowadays, various multiplex reverse transcription assays have been developed (13), but until now, there has not been enough study on SYBR Green multiplex real-time PCR for detection of respiratory viruses, a technique shown to be fast and cost effective.
2. Objectives
The aim of this study was recognizing the panel of respiratory RNA viruses in hospitalized patients of all age groups during different seasons in one year by SYBR Green multiplex real-time PCR assay.
3. Materials and Methods
2.1. Study Participants
Randomized 172 respiratory specimens of hospitalized patients with influenza-like illnesses (ILI) of all age groups, negative for influenza viruses, were collected from 23 September 2012 until 22 September 2013 from National Influenza Center, school of Public Health, Tehran University of Medical Sciences.
2.2. Laboratory Procedures
RNA was extracted from 200 µL of each sample using high pure viral nucleic acid kit (Roche, Germany). For synthesis of cDNA, 16 µL master mix (Fermentas, Germany) consisted of RT-PCR buffer, dNTPs, random hexamer, reverse transcriptase enzyme and RNase inhibitor was mixed with 24 µL of extracted RNA. Reverse transcription was performed at 37°C for one hour. Three multiplex SYBR Green real-time PCR assays were developed for simultaneous detection of 12 respiratory RNA viruses. Each multiplex set was designed for detection of four viruses as follows: the first set was respiratory syncytial virus (RSV), human metapneumovirus (HMPV), rhinovirus (RV), and enterovirus (EV); the second set was parainfluenza viruses 1-4 (PIV1-4) and the third set was coronaviruses NL63, 229E, SARS, OC43. The next step was primer designing and the most important factor was the Tm of PCR products which must be different in each primer pair for specific detection of each virus according to the melt curve. Table 1 shows the properties of the primers.
| Primer Name | Primer Sequence (5’-3’) | Target Gene | Product Length | Product Tm |
|---|---|---|---|---|
| OC43-F | CGATGAGGCTATTCCGACTAGGT | S | 76 | 89.26 |
| OC43-R | CCTTCCTGAGCCTTCAATATAGTAACC | |||
| SARS-F | AAGCCTCGCCCAAAAACGTAC | NC | 229 | 99.12 |
| SARS-R | AAGTCAGCCATGTTCCCGAA | |||
| NL63-F | ACGTACTTCTATTATGAAGCATGATATTAA | 1a | 103 | 81.29 |
| NL63-R | AGCAGATCTAATGTTATACTTAAAACTACG | |||
| 229E-F | CATACTATCAACCCATTCAACAAG | M | 137 | 94.47 |
| 229E-R | CACGGCAACTGTCATGTATT | |||
| PIV1-F | ACCTACAAGGCAACAACATC | HN | 129 | 87.75 |
| PIV1-R | CTTCCTGCTGGTGTGTTAAT | |||
| PIV2-F | CCATTTACCTAAGTGATGGAA | HN | 116 | 89.04 |
| PIV2-R | CGTGGCATAATCTTCTTTTT | |||
| PIV3-F | ACCAGGAAACTATGCTGCAGAACGGC | NP | 234 | 94.45 |
| PIV3-R | GATCCACTGTGTCACCGCTCAATACC | |||
| PIV4 -F | CAAATGATCCACAGCAAAGATTC | NP | 123 | 92.75 |
| PIV4-R | ATGTGGCCTGTAAGGAAAGCA | |||
| EV-F | GGCCCCTGAATGCGGCTAATCC | NCR | 151 | 98.76 |
| EV-R | GCGATTGTCACCATAAGCAGTCA | |||
| RV-F | GGTGTGAAGACTCGCATGTGCT | NCR | 277 | 93.27 |
| RV-R | CCAAAGTAGTCGGTTCCGCTTCTGA | |||
| RSV-F | GGAACAAGTTGTTGAGGTTTATGAATATGC | F | 139 | 96.37 |
| RSV-R | TTCTGCTGTCAAGTCTAGTACACTGTAGT | |||
| HMPV-F | AACCGTGTACTAAGTGATGCACTC | NP | 212 | 90.98 |
| HMPV-R | CATTGTTTGACCGGCCCCATAA |
a Abbreviations: F, fusion; HN, hemagglutinin neuraminidase; M, matrix; NC, nucleocapsid; NCR, non-coding region;. NP, nucleoprotein; S, spike.
In all the sets, cDNA was amplified by a real-time PCR using Power SYBR Green PCR Master Mix Kit (ABI, USA) in OneStep ABI instrument (ABI, USA). Each reaction had a total volume of 25 µL, including 12.5 µL SYBR Green master mix, 200 nmol of each forward and reverse primers, 5 µL cDNA plus 7.1 µL ddH2O. The cycling conditions included an initial denaturation step of 10 minutes at 94°C, followed by 40 cycles of 15 seconds at 95°C, one minute at 55°C and one minute at 60°C. Fluorescent detection was at the end of each cycle. Melting curve analysis program was used for identification of specific PCR products. After the last cycle, the temperature was increased to 94°C, then decreased to 75°C and slowly increased to 94°C at a rate of 0.1°C per second, with continuous fluorescence monitoring. Positive and negative controls were added in each set. To prevent contamination, RNA samples and PCR master mixes were prepared under biosafety hoods in different rooms.
4. Results
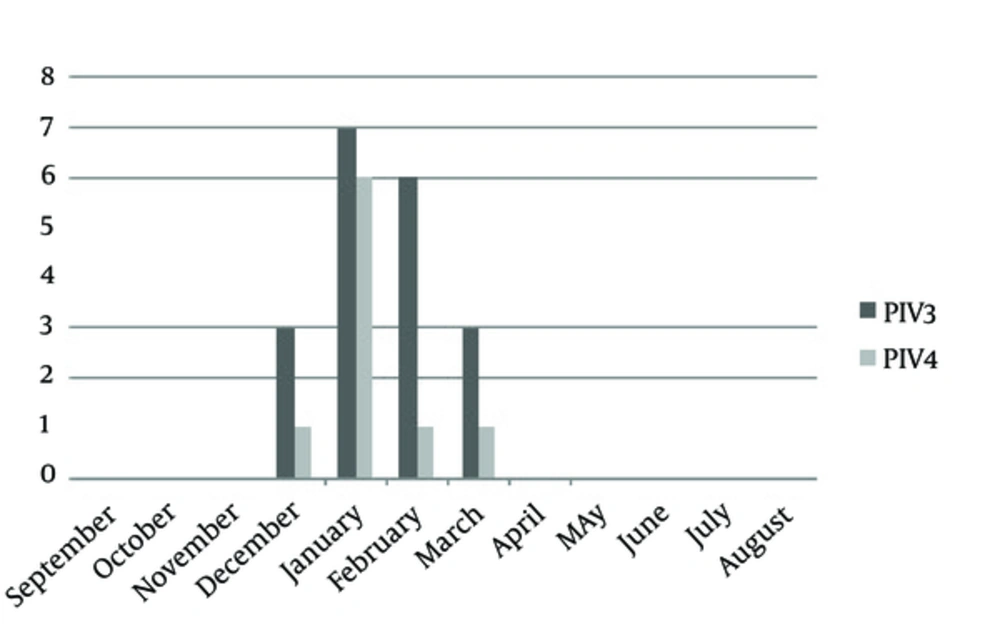
During the September 2012 through September 2013, 3276 throat swab specimens were collected from hospitalized patients of all ages with symptoms of ILI. Initially, all the specimens were tested for influenza viruses by superscript III platinum one-step quantitative real-time PCR assay (Invitrogen, USA), selective for the matrix gene; 2514 samples were negative for influenza viruses. Of which 172 samples were randomly selected for this study. Of 172 specimens, the male:female proportion was 96:76. The mean age was 33.63 years and the median was 31 years. Of 172 samples, in the first set we did not detect any RSV, RV, EV or HMPV. In the second set, there was no PIV1 and PIV2, but 19 (11.04%) PIV3 and 9 (5.23%) PIV4 were detected. In the set of coronaviruses NL63, 229E, SARS, OC43, the only detected virus was 1 (0.58%) NL63. This was the first time that human corona virus NL63 was detected in Iran. All the PIV3- and PIV4-positive samples were detected during December to March (Figure 1) and one corona virus NL63 belonged to a 28-day-old newborn in February. Among positive samples, the male:female proportion was 20:9, indicating that males were more affected than females. The mean age was 33.27 years and the median was 32 years. The standard curve and melt curves for positive samples are shown in Figures 2 and 3.
5. Discussion
In this study, we described a multiplex SYBR Green real-time PCR assay for detection of 12 respiratory RNA viruses, belonging to three families of viruses: Picornaviridae (RV and EV), Paramyxoviridae (RSV, HMPV, hPIV 1-4) and Coronaviridae (coronaviruses 229E, SARS, NL63, OC43). Enteroviruses and RVs can be detected in 25 - 30% of respiratory tract infections. Nearly 80% of all children have experienced RV infections by the age of two years. Human EV type 68 has been identified in respiratory tract specimens and has been associated with respiratory diseases (14). RSV is the most important cause of severe LRI in infants and young children worldwide. In temperate climates, RSV has been significantly documented as a winter epidemics agent of acute LRI (bronchiolitis and pneumonia) (2). Some studies have reported difficulties in detection of RSV with swabs. RSV lability was suggested as a reason of decreasing the sensitivity of detection (15). HMPV is the cause of 5 - 7% of viral respiratory tract infections in hospitalized children (16). HMPV outbreaks usually occur at the same time with the RSV season (17, 18). According to the phylogenetic analysis, HMPV is the closest human virus to RSV (19).
Parainfluenza viruses cause a range of respiratory illness from 30 - 50% of URIs including otitis media to 0.3% of hospitalized children. Croup is one clinical sign of infection with parainfluenza viruses. HPIV1 and HPIV3 are the important causes of upper and lower respiratory tract infections in infants, young children and immunocompromised people (20). Human respiratory coronaviruses infect all age groups. Nearly 10% of all URI and LRI in children are caused by these viruses (21). Corona viruses are often co-detected with other respiratory viruses, especially RSV (22), but in this study, we did not have co-infections. Respiratory disease can be caused by one of the known respiratory viruses. Nowadays, there are different methods for detection of viruses which may lead to different results. The use of some diagnostic methods such as cell culture might have long delays before the final results. Sensitive and rapid detection of viral infections is essential for reducing the nosocomial transmission and limiting the overuse of antibiotics (15); molecular detection methods have been widely used.
It is notable that singleplex PCR is too arduous, expensive and sample consuming. Multiplex PCR approaches were becoming more and more acceptable for the detection of respiratory viruses, since it is cost- and time-effective (5). Multiplex conventional PCR has false positive results due to contamination and requires further precautions, more time and two separate machines and facilities (5). Multiplex Taqman real-time PCR is much more expensive than the other methods. In this study, we developed a multiplex SYBR green real-time PCR for detection of 12 RNA respiratory viruses. There were 39 (22.6%) children less than six years old, in which 1 (2.56%) coronavirus NL63 was detected. Esper et al. (23) identified an incidence of 8.8% out of 895 pediatric samples positive for coronavirus NL63 annually and a French study found an incidence of 9.3% out of 300 specimens during five months (24).
One of the reasons for different results in detection of respiratory RNA viruses is using different clinical criteria for selection of patients in different studies. In this study, RV and EVs were not detected, which might have been due to the selection criteria, because we selected the patients hospitalized with respiratory infections, while we knew that RV and EV usually cause mild respiratory infections. We did not detect RSV which is a relatively labile virus and might be lost by freezing and defreezing of the samples. Other limitations of our study were the small sample size and wide age range. The ability of SYBR green real-time PCR in detection of several viruses was assessed and the assay was found easy to use and cost- and time-effective, but for better understanding of sensitivity and specificity, it should be compared with other methods.