1. Background
Multidrug-resistant (MDR) Acinetobacter baumannii isolates have emerged as the causes of nosocomial infections worldwide. Recently, A. baumannii isolates have proved almost always extensively drug-resistant (XDR), which is defined by resistance to all antibacterial agents except tigecycline and colistin. The treatment of XDR A. baumannii infections is a major challenge given the lack of effective treatment options and limited management experience. Unfortunately, the increased use of tigecycline and colistin as salvage therapy has been associated with the emergence of pan-drug resistance, against which no known antibacterial agents retain activity (1). There is a huge risk of these “superbugs” extending into the community and threatening public health (2). Acinetobacter is an important opportunistic pathogen of hospital-acquired infections, particularly in intensive care units, and as a leading cause of morbidity and mortality among hospitalized burn patients. Indeed, A. baumannii is usually responsible for up to 10% of hospital-acquired infections and increases mortality up to 70% (3). Acinetobacter is a causative agent of nosocomial bacteremia, pneumonia, urinary tract infection, and meningitis (4, 5).
A wide array of antibacterial resistance mechanisms has been reported for resistant A. baumannii strains (6) such as efflux pumps, outer membrane proteins, and Β-lactamases (7). Nevertheless, it has been established that efflux mechanisms play an important role in multidrug resistance in Gram-negative bacteria, particularly in A. baumannii isolates (8). Efflux pumps are composed of transport proteins that pump out a broad range of toxic substrates such as biocides and antibiotics from bacteria, in an energy-dependent manner (9). It has recently been reported that resistance to Β-lactams, aminoglycosides, chloramphenicol, tetracycline, erythromycin, and the dye ethidium bromide in clinical isolates are due to the overexpression of the AdeABC pump (9, 10). The AdeABC, a kind of efflux pump system initially detected in an MDR isolate of A. baumannii, is responsible for increased resistance to different antibacterials (8). The chromosomally encoded AdeABC is a tripartite efflux machinery that belongs to the RND-type family. The AdeC is a homologous outer membrane protein (OprM) from Pseudomonasaeruginosa. The AdeA is most similar to membrane fusion proteins, whereas the AdeB contains twelve transmembrane segments and exhibits a high degree of identity with several RND proteins.
The structural genes adeA, adeB, and adeC are contiguous and directly oriented, suggesting that they constitute an operon (10). The overexpression of efflux pumps in A. baumannii is a common mechanism of multidrug resistance in this nosocomial pathogen. An increased efflux pump expression is often assumed from the minimum inhibitory concentrations (MICs) of dyes and antibiotics, without measurement of efflux levels (11). The active efflux of antibacterial agents is a mechanism in A. baumannii isolates whereby they can become MDR. A combined use of efflux pump inhibitors such as phenylalanine-arginine Β-naphthylamide (PAΒN, also called MC-207, 110) with pump substrates is under exploration to overcome efflux-mediated multidrug resistance. The PAΒN is one of the best-studied efflux pump inhibitors. The PAΒN was originally detected in 1999 and characterized further in 2001 as a broad-spectrum efflux pump inhibitor, capable of significantly reducing antibiotic resistance in P. aeruginosa (12). Accordingly, the aim of this study was to analyze the contribution of the active efflux system to imipenem resistance in the clinical isolates of A.baumannii using the efflux pump inhibitor, the PAΒN.
2. Objectives
The aims of this study were to determine the frequency of the AdeABC genes and the role of efflux pump (s) in the imipenem resistance of A. baumannii strains isolated from burn patients.
3. Materials and Methods
3.1. Bacterial Identification
From June to October 2013, a total of 60 non-duplicate non-consecutive isolates of A. baumannii were recovered from 240 wound samples of burn patients admitted to the burn unit of Shahid Motahari Burn Hospital, Tehran, Iran. Wound exudates were collected by swabbing and immediately transported to the microbiology laboratory of the department of microbiology of Shahid Beheshti university of medical sciences, Tehran, Iran. According to conventional biochemical tests (13), the typical reaction of A. baumannii is positive to glucose and negative to oxidase, maltose, indole, mannitol, esculin, and H2S. A. baumannii also has ALK/ALK reaction on triple sugar iron agar. Standard identification, confirmation, and complete method were conducted using the Microgen Identification kit (Microgen TM, UK). Samples confirmed as an A. baumannii were stored in Tryptic Soy Broth (TSB) (Merck, Germany), containing glycerol (20%) at -70°C (14).
3.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility test to tetracycline (TE, 10 μg), meropenem (MEM: 10 μg), amikacin (30 μg), imipenem (10 μg), cefotaxime (30 μg), ceftriaxone (30 μg), piperacillin/tazobactam (100/10 μg), colistin sulfate (10 μg), piperacillin (100 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), cefepime (30 μg), trimethoprim-sulfamethoxazole (2.5 μg), and gentamicin (10 μg) (Mast, UK) was performed using the Kirby-Bauer disk diffusion method based on the clinical laboratory standards institute (CLSI) 2012 guidelines (15). Briefly, a bacterial suspension was obtained from overnight cultures. The turbidity of each bacterial suspension was adjusted equivalent to a no. 0.5 McFarland standard and then inoculated on Müller-Hinton agar (Merck, Germany). The diameter of the inhibition zones was measured after incubation at 35ºC for 18 - 24 hours, and the data were reported as susceptible, intermediate, and resistant. Escherichia coli ATCC 25922 was used as a quality control strain.
3.3. Minimum Inhibitory Concentration (MIC)
Strains which were resistant to imipenem and ceftazidime during the disk diffusion test were re-checked and their MICs were determined via the broth microdilution method in accordance with the 2012 guidelines of the CLSI (15). Escherichia coli ATCC 25922 was used as a quality control strain. Antibiotic powders were dissolved in an appropriate solvent or sterile deionized water according to the manufacturer’s recommendations. The test concentrations for the antibiotics were 256 µg/mL, 128 µg/mL, 64 µg/mL, 32 µg/mL, 16 µg/mL, 8 µg/mL, 4 µg/mL, 2 µg/mL, 1 µg/mL, 0.5 µg/mL, and 0.25 µg/mL. Each well of a 96-well microtiter plate (Extra Gene-Company-product No.EL-1190-FCS) contained a total volume of 100 µL of the antibiotic dilution and Müller-Hinton broth. Subsequently, the 0.5 McFarland suspension was diluted 1: 20 to yield 5 × 106 CFU/mL. When 0.01 mL of this suspension was inoculated into the broth, the final test concentration of the bacteria was approximately 5 × 105 CFU/mL. The correct density of the turbidity standard was verified by measuring absorbance using a spectrophotometer. The absorbance at 625 nm should be 0.08 to 0.13 for the 0.5 McFarland standard. The samples were incubated at 37°C for 18 to 24 hours. The lowest concentration of the antibiotics that did not have visible bacterial growth was defined as the MIC (15).
3.4. Treatment of the Efflux Pump Inhibitor
Changing in susceptibility to imipenem in the presence of 100 µg/mL of the PAΒN (Sigma-Aldrich, Oakville, ON, Canada) was tested as described under “Susceptibility Testing” above. Specifically, susceptibility to imipenem was tested in the presence or absence of the PAΒN in parallel. Following the addition of imipenem and the bacterial cell inoculum, 2 μL of the 5 mg/mL stock of PAΒN was added to the microplate wells. (total volume, 100 µL). The rest of the procedures were carried out as described above (16). To check the acridity of the test and evaluate the effect of the PAΒN on the bacterial grow, all the bacteria were cultured in the Mueller Hinton broth, containing the PAΒN (100 µL/mL).
3.5. DNA Extraction
In this stage, 2 ml of 24 hours grown A. baumannii cultures in a brain heart infusion (BHI) medium (Merck, Germany) were transferred into Eppendorf microfuge tubes and centrifuged at 8000 rpm for 10 min. The pellets were dissolved in the lysis buffer (700 µL) (NaCl 1 M, Tris -HCl 1 M, EDTA 0.5 M and DD/water). Next, 20 µL of the Sodium Dodecyl sulfate (SDS) solution (25%) and 3 µL of proteinase K (20 mg/mL) were added to the mixture. The microfuge tubes were incubated at 60°C for 1 h. After the lysis of the bacterial cells, 620 µL of phenol, chloroform, and isoamylalcohol (25: 24: 1 Vol/ Vol) was added to the above solution and carefully shaked and the mixtures were centrifuged at 10000 rpm for 10 min at 4°C. The supernatant was transferred to another tube and 2 mL of cold ethanol (75%) was added till 1 h. Then the DNA was precipitated at 10,000 rpm for 10 minutes. The supernatant was discarded, and the DNA was dissolved in a Tris-EDTA (TE) buffer (50 µL), containing RNase (15 µL). The extracted DNA samples were kept at -20°C for further investigation.
3.6. Detection of the AdeABC Genes by Polymerase Chain Reaction
PCR was performed on the A. baumannii isolates for the AdeABC genes. The primers utilized are presented in Table 1. Briefly, the 25 µL PCR mixture contained 2.5 µL of bacterial DNA, 10 pM of each primer, 1.5 mM of MgCl2, 250 µM of each dNTP, 10 Mm of Tris-HCL (pH: 9.0), 30 Mm of KCL, and 1 U of Taq DNA polymerase (Bioneer Company, Korea, Cat. No. K-2012). Reactions were performed on a thermal cycler (Eppendorf, Master Cycler Gradient). Amplification was carried out with the following thermal cycling conditions: 5 min at 94°C and 30 cycles of amplification consisting of 1 min at 94°C, 1 minute at 45°C - 56°C, and 1 minute at 72°C, with 5 minutes at 72°C for the final extension. The PCR product bands were analyzed after electrophoresis on agarose gel (1%) at 90 V for 50 minutes in 1X boric acid (TBE), containing ethidium bromide, and the result was checked under UV irradiation. A PCR purification kit (Bioneer Co., Korea) was used to purify the PCR products, and sequencing was performed by the Bioneer Company (Korea). The nucleotide sequences were analyzed with the Chromas 1.45 software and BLAST in NCBI.
3.7. Statistical Analysis
This research was a descriptive-application study. MINITAB16 software was used for the statistical analyses. The P value and confidence intervals were less than 0.05 and 95%, respectively.
3.8. Nucleotide Sequence Accession Numbers
The nucleotide sequences of the adeA, adeB, and adeC were submitted to the GenBank database and were assigned accession numbers KJ711551, KJ711555, and KJ711554, respectively.
4. Results
4.1. Bacterial Isolates and Antibiotic Resistance
Totally, 60 A. baumannii strains were isolated from 240 wound samples of burn patients admitted to the burn unit at Shahid Motahari Burn Hospital (Tehran, Iran). Fifteen (25 %) strains were isolated from female patients and 45 (75 %) from male patients. The age range of the patients was between 7 and 65 years. The resistance of A. baumannii isolates to the tested antibiotics was 60 (100%) to cefotaxime, ceftazidime, ceftriaxone, ciprofloxacin, cefepime, piperacillin, meropenem, co-trimoxazole, and piperacillin tazobactam; 58 (97%) to imipenem; 56 (94%) to gentamicin; 50 (81%) to amikacin; 45 (76%) to tetracycline; and 0 (0.0%) to colistin (Table 2). The MICs of ceftazidime and imipenem for the A. baumannii isolates are presented in Table 3.
| Antibiotic | Resistant | Intermediate | Sensitive |
|---|---|---|---|
| Gentamicin | 56 (94) | 0 (0) | 4 (6) |
| Co-trimoxazole | 60 (100) | 0 (0) | 0 (0) |
| Amikacin | 50 (83) | 5 (8) | 5 (8) |
| Imipenem | 58 (97) | 2 (3) | 0 (0) |
| Cefotaxime | 60 (100) | 0 (0) | 0 (0) |
| Cefepime | 60 (100) | 0 (0) | 0 (0) |
| Meropenem | 60 (100) | 0 (0) | 0 (0) |
| Ciprofloxacin | 60 (100) | 0 (0) | 0 (0) |
| Ceftriaxone | 60 (100) | 0 (0) | 0 (0) |
| Piperacillin/tazobactam | 60 (100) | 0 (0) | 0 (0) |
| Ceftazidime | 60 (100) | 0 (0) | 0 (0) |
| Tetracycline | 45 (76) | 7 (11) | 8 (13) |
| Piperacillin | 60 (100) | 0 (0) | 0 (0) |
| Colistin | 0 (0) | 5 (5) | 55 (95) |
a Data are presented as No. (%).
| Antibiotics | MIC a | ||
|---|---|---|---|
| MIC50 | MIC90 | MIC Range | |
| Imipenem | 64 | 128 | 16 - 128 |
| Ceftazidime | 256 | > 256 | 32 - > 256 |
aData are presented as µg/ml.
4.2. Effects of the Efflux Pump Inhibitor on Imipenem Resistance
To determinate the role of the efflux pump in the imipenem-resistant phenotype of the 60 A. baumannii isolates, we evaluated the variation of the MIC of imipenem in the presence or absence of the PAΒN (100 µg/mL). Our comparison analysis indicated that in the presence of the pump inhibitor, the quantity of the MICs of imipenem in most of the isolates decreased in the presence of the efflux pump inhibitor (Table 4). Based on a fourfold or greater decrease in the MICs as the criterion for significance (16), the MICs for 58 of the 60 isolates (96.6%) were reduced significantly by 4- to 64-fold. In addition, a twofold decrease was detected in the 2 (3.3%) remaining isolates in the presence of the efflux pump inhibitor. All the grown bacteria cultured in the Mueller-Hinton broth, containing the PAΒN (100 µg/mL) without with no any imipenem, showed well growth of bacteria, indicating that the PAΒN (100 µg/mL) did not have an antibacterial effect itself. These results showed that drug efflux systems were associated with resistance to imipenem in the A. baumannii isolates in the present study.
a Data are presented as No. (%).
4.3. Analysis of the AdeABC Genes
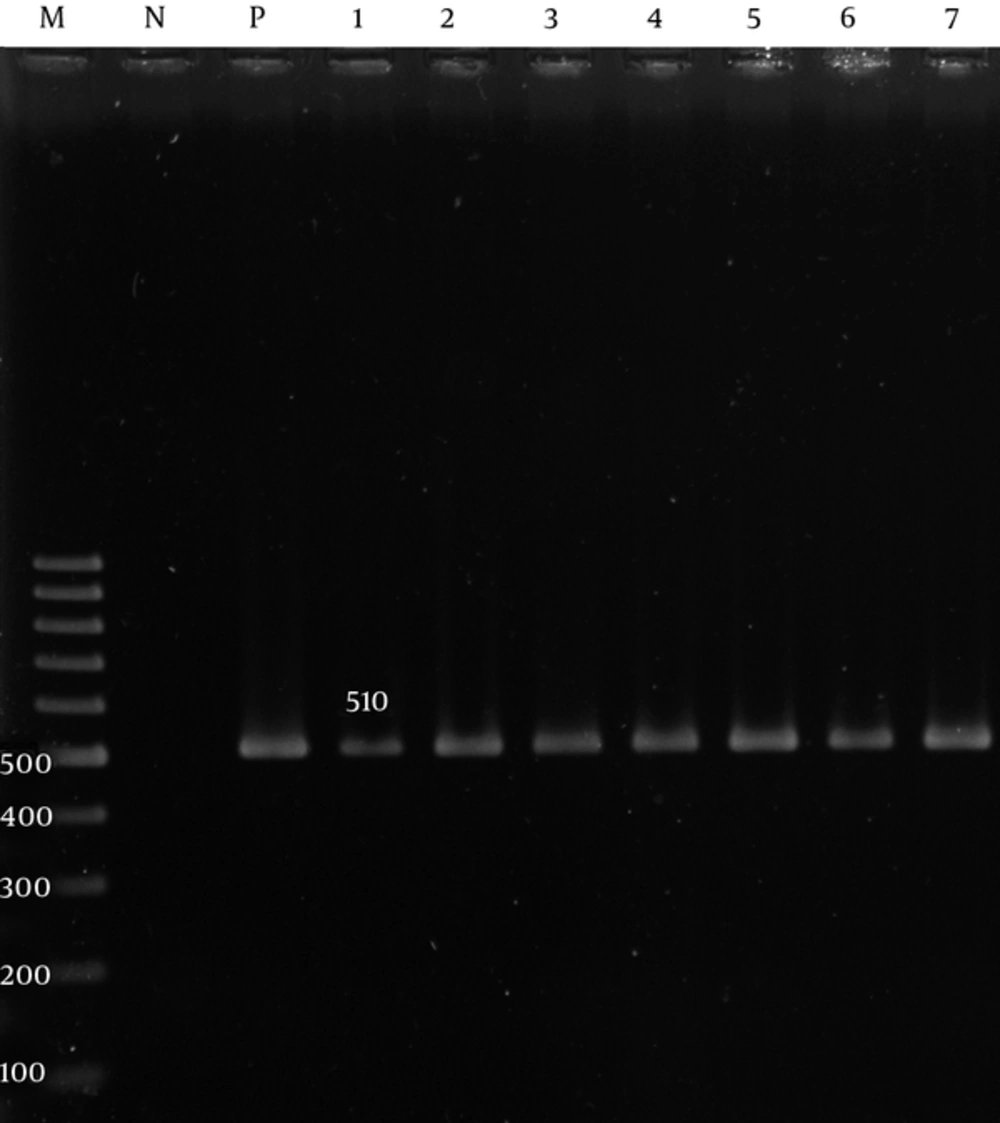
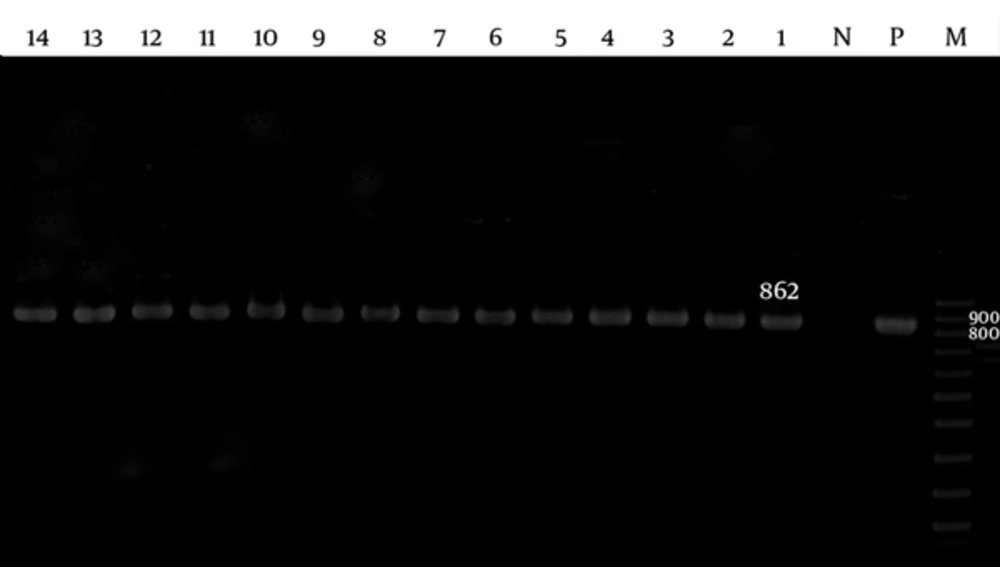
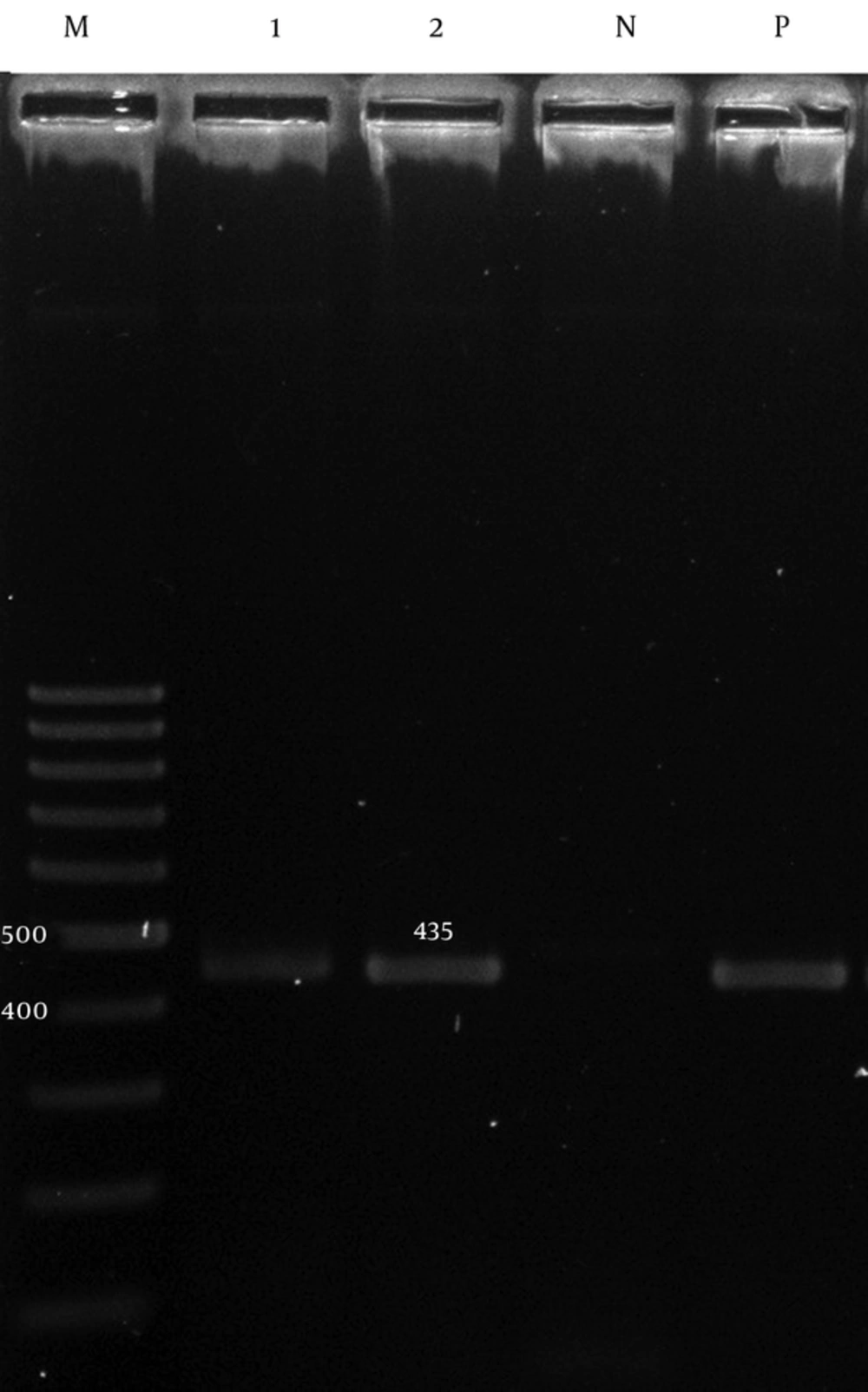
The adeA and adeB were detected in 60 (100%) of the isolates (Figures 1 and 2), while the adeC was detected in 51 (85%) isolates (Figure 3).
4.4. Mortality Rate
The mortality rate due to A. baumannii infection was 5 (8.3%) among the hospitalized burn patients.
5. Discussion
Infection can lead to the deterioration of the burn healing process and severe systemic complications and is the leading cause of mortality and morbidity in patients (19). A. baumannii is an opportunistic pathogen that is particularly successful at colonizing and persisting in the hospital environment. It is also able to resist desiccation and survive on inanimate surfaces for months (4). The isolation of A. baumannii strains from burn centers is frequent and has been reported by various global studies, including one from Iran (20). In the present study, the resistance rates of the isolates were as follows: 60 (100%) to co-trimoxazole, cefotaxime, cefepime, meropenem, ciprofloxacin, ceftriaxone, piperacillin/Tazobactam, ceftazidime, and piperacillin; 58 (97%) to imipenem; 56 (94%) to gentamicin; 50 (81%) to amikacin; 45 (76%) to tetracycline; and 0 (0%) to colistin. Accordingly, the best coverage against the study isolates was obtained with colistin. Various studies have clearly demonstrated that the incidence of resistant A. baumannii strains is on the increase worldwide (20).
These studies have also shown that the resistance of these strains is not only against Β-lactamases, including carbapenemas and the third generation of cephalosporins, but also against other drug divisions such as fluoroquinolones and aminoglycosides. The other importance of these bacteria is related to their multi-drug resistance, which restricts their treatment. Mohajeri et al. reported that all the isolates in their study were resistant to imipenem and meropenem as well as to other antimicrobial agents (21). Shahcheraghi et al. (2011) determined that 12% of the A. baumannii isolates in their investigation were resistant to colistin (13). Fallah et al. reported that the resistance rate of the A. baumannii strains to colistin in their research was 2 (1.8%) (20). Therefore, colistin can be helpful in treating A. baumannii-related infections in burn patients. Efflux pump systems are an extremely important cause of multi-drug resistance (22). Although high level resistance may not occur as a result of multidrug efflux pumps alone, the overexpression association of these genes among MDR isolates cannot be ignored. Efflux pump inhibitors have been shown to reverse multidrug resistance in A. baumannii and other bacteria (23).
The effects of these compounds such as the PAΒN on the antimicrobial susceptibility have been examined in some studies. Valentine et al. found that the addition of the PAΒN at a final concentration of 100 µg/ml greatly reduced the MIC of ciprofloxacin from 2- to 8-fold (16). Pan Hou et al. noted that after exposure to the efflux pump inhibitor, the PAΒN, a 4- to 32-fold reduction in the MICs of imipenem was observed in 33 (66%) isolates of imipenem-resistant A. baumannii (24). Szabo´ et al. reported that the addition of the PaΒN at different concentrations reduced the MICs of various antibiotics (17). We observed that the imipenem susceptibility of most of the isolates was increased in the presence of the PAΒN by 4- to 64-fold. The results suggest that multi-drug efflux pumps play a role in the mechanism of the resistance in A. baumannii strains. It has recently been reported that resistance to antibiotics is due to the overexpression of the AdeABC pump (10). In the current study, the adeA and adeB were detected in 60 (100%) of the isolates, while the adeC was detected in 51 (85%) isolates. The AdeABC operon is present in 80% (from 53% to 97%) of A. baumannii strains (25). The mortality rate in the present study was 8.3%. Vahdani et al. reported that the mortality rate caused by the A. baumannii strains in their study was 26%. Other researchers have reported a wide range of mortality from 7% to 43% (26).
The results of the present study indicate that the antibiotic efflux pump has a role in conferring resistance to antibiotics in A. baumannii isolates. Thus, efforts should be aimed at detecting such resistant A. baumannii along with their resistance mechanisms. Moreover, further research is needed to find more suitable and effective therapies against A. baumannii isolates and since the available data support the development of efflux pump inhibitors for use in combination with antibiotics, it is advisable that the cytotoxicity and mutagenicity of inhibitors in future studies be evaluated.