1. Background
CTX-M-type enzymes are a group of non-TEM (temoniera) and SHV (sulfhydryl-variable) class A extended-spectrum β-lactamases (ESBLs) with rapid spread ability amongst Gram-negative bacteria (1). CTX-M-producing Klebsiella pneumoniae are becoming increasingly prevalent in clinical and nosocomial environments (2). CTX-M-type ESBLs-producing strains were documented for the first time in the late 1980s in Japan, Europe and Argentina (1). During the past two decades, CTX-M-type enzymes have been documented as the most prevalent ESBL enzymes, especially amongst ESBL-producing Enterobacteriaceae isolated from clinical specimens in Europe, Asia and South America (3). In Taiwan and South Korea, the rates of CTX-M production were found to be 58.5% and 32.7% amongst ESBL-producing isolates of K. pneumoniae, respectively (1). In another recent study from Iran, 26.9% was documented as the rate of CTX-M-1 ESBL-production amongst K. pneumoniae isolates (4). One of the most important problems associated with ESBL-producing bacterial isolates is believed to be nosocomial outbreaks of K. pneumonia (5).
To date, more than 50 different β-lactamases have been identified, which are grouped on the basis of amino acid sequences into five subtypes including CTX-M-1 (CTX-M-1, -3, -10, -11, -12, -15, -28 and FEC-1), CTX-M-2 (CTX-M-2, -4, -5, -6, -7, -20 and TOHO-1), CTX-M-8 (CTX-M-8), CTX-M-9 (CTX-M-9, -13, -14,-16, -17, -19, -21, -24, -27 and TOHO-2) and CTX-M-25 (6). Recently, although two more additional clusters have been reported, the rapid spread of CTX-M-type enzymes is facilitated by incorporation of corresponding resistance genes in mobile genetic elements such as plasmids, transposons and integrons (7, 8). Global dissemination of blaCTX-M genes has been documented, and CTX-M enzymes have been identified in hospitalized and out patients all around the world (9). This rapid dissemination of CTX-Ms all over the world has been described as the “CTX-M pandemic” (7).
2. Objectives
Little is known about the different types of CTX-M ESBLs among Klebsiella isolates in Iran. Therefore this study aimed to determine types of CTX-M ESBLs among ESBL producing Klebsiella isolates from patients in Kashan, Iran.
3. Materials and Methods
3.1. Bacterial Isolates
During December 2012 to November 2013, 100 Klebsiella isolates were collected from hospitalized patients of both sexes (64% female and 36% male) at Shahid Beheshti Hospital of Kashan, Iran. The isolates were recovered from urine (n = 75), tracheal aspirate (n = 12), sputum (n = 7) and blood (n = 6). The Klebsiella strains were identified by standard microbiological tests (10). All strains were stored at -70°C in Tryptic Soy Broth (TSB) medium supplemented with 10% glycerol for further studies.
3.2. Antibiotic Susceptibility Testing and Confirmation of Extended-Spectrum β-Lactamases Production
Antimicrobial susceptibility testing was performed by the disk diffusion method according to the clinical and laboratory standards institute (CLSI) guidelines (11), and fourteen antibiotic disks were used including: ampicillin (30 Μg), aztreonam (30 Μg), amoxicillin/clavulanic acid (20 Μg), cephalothin (30 Μg), cefixime (30 Μg), nalidixicacid (30 Μg), trimethoprim-sulfamethoxazole (25 Μg), imipenem (10 Μg), ceftazidime (30 Μg), cefoxitin (30 Μg), cefteriaxon (30 Μg), gentamicin (10 Μg), ciprofloxacin (5 Μg) and nitrofurantoin (300 Μg) (Mast Companies, UK). The quality control organism was Escherichia coli ATCC 25922. The ESBL production of Klebsiella isolates was confirmed by the double disk synergy test (DDST) using disks of ceftazidime (30 Μg) and cefotaxime (30 Μg) with and without clavulanic acid (10 Μg) (CLSI, 2012). The K. pneumonia ATCC 700603 strain was used as the positive control and E. coli strain ATCC 25922 was used as the negative control (11).
3.3. Genotype Detection
DNA of ESBL-producing Klebsiella isolates was extracted using the boiling method. The Klebsiella strains were cultured in Luria Bertani (LB) broth at 37°C for 18 hours. Next, bacteria in 1.5 mL of LB broth were pelleted and suspended in 250 Μl of sterile deionized water and incubated at 100°C for 10 minutes. After centrifugation, the supernatant was used as a template DNA for the Polymerase Chain Reaction (PCR) assay.
Polymerase Chain Reaction amplification was carried out and specific primers (Table 1) were used for diverse CTX-M types (CTX-M-1, CTX-M-2, CTX-M-8 and CTX-M-9) (12). Amplification was performed in a total volume of 25 ΜL, including 1.5 ΜL of template DNA, 2.5 ΜL of 10 X reaction buffer, 10 pM of each of the forward and reverse primers, 200 Μm dNTP, 1.5 Μm MgCl2, and 1U Taq DNA polymerase (CinnaGen, Tehran, Iran). The amplification reaction was carried out in a Thermal Cycler (Eppendorf master cycler®, MA) with an initial denaturation (94°C for five minutes) followed by 30 cycles of denaturation (94°C for 25 seconds) annealing (52°C for 40 seconds), and extension (72°C for 50 seconds) and a single final extension at 72°C for six minutes (12). Clinical strains with defined CTX-M β-lactamases (CTX-M-1, CTX-M-2, CTX-M-8 and CTX-M-9 groups) were used as the positive control. The amplified products were electrophoresed on 2% agarose gel and visualized on a gel document system (Biorad, UK) after staining with ethidium bromide (0.5 mg/mL).
| Gene | Primer | Sequence (5’-3’) | Amplification Product (bp) | Reference |
|---|---|---|---|---|
| blaCTX-M | CTX-MA | CGCTTTGCGATGTGCAG | 590 | 10 |
| CTX-MB | ACCGCGATATCGTTGGT | |||
| blaCTX-M-1 | M-1F | AAAAATCACTGCGCCAGTTC | 415 | 10 |
| M-1R | AGCTTATTCATCGCCACGTT | |||
| blaCTX-M-2 | M-2 | CGACGCTACCCCTGCTAT | 552 | 10 |
| M-2R | CCAGCGTCAGATTTTTCAGG | |||
| blaCTX-M-8 | M-8F | ACGCTCAACACCGCGATC | 490 | 10 |
| M-8R | CGTGGGTTCTCGGGGATAA | |||
| blaCTX-M-9 | M-9F | CAAAGAGAGTGCAACGGATG | 205 | 10 |
| M-9R | ATTGGAAAGCGTTCATCACC |
3.4. DNA Sequencing
The purified PCR products were sequenced using the ABI Capillary System (Macrogen Research, Seoul, Korea). Sequences were compared using the online BLAST software (http://www.ncbi.nlm. nih.gov/BLAST/).
4. Results
The antibiotic resistance pattern of Klebsiella strains isolated from different clinical specimens using the disk diffusion method is shown in Table 2.
| Antibiotic | Urine (N = 75), % | Tracheal Aspirate (N = 12), % | Sputum (N = 7), % | Blood (N = 6), % | Total (N = 100) |
|---|---|---|---|---|---|
| Ampicillin | 69 (92) | 11 (91.7) | 7 (100) | 5 (83.3) | 92 (92) |
| Nalidixic Acid | 38 (50.7) | 10 (83.3) | 1 (14.3) | 2 (33.3) | 51 (51) |
| Cotrimoxazole | 16 (21.3) | 3 (25) | 2 (28.6) | 0 (0) | 21 (21) |
| Ciprofloxacin | 23 (30.7) | 8 (66.7) | 1 (14.3) | 1 (16.7) | 33 (33) |
| Ceftriaxone | 30 (40) | 3 (25) | 1 (14.3) | 1 (16.7) | 35 (35) |
| Aztreonam | 27 (36) | 7 (58.3) | 1 (14.3) | 1 (16.7) | 36 (36) |
| Ceftazidime | 26 (34.7) | 6 (50) | 1 (14.3) | 0 (0) | 33 (33) |
| Cephalothin | 32 (42.7) | 9 (75) | 1 (14.3) | 2 (33.3) | 44 (44) |
| Gentamicin | 11 (14.7) | 3 (25) | 1 (14.3) | 0 (0) | 15 (15) |
| Nitrofurantoin | 32 (42.7) | 5 (41.7) | 1 (14.3) | 2 (33.3) | 40 (40) |
| Amoxicillin-Clavulanic Acid | 42 (56) | 5 (41.7) | 2 (28.6) | 0 (0) | 49 (49) |
| Cefoxitin | 20 (26.7) | 5 (41.7) | 1 (14.3) | 0 (0) | 26 (26) |
| Cefotaxime | 48 (64) | 8 (66.7) | 2 (28.6) | 1 (16.7) | 59 (59) |
| Imipenem | 5 (6.7) | 2 (16.7) | 0 (0) | 0 (0) | 7 (7) |
The highest and lowest resistance rates were found for ampicillin (92%) and imipenem (7%), respectively. The highest resistance rate to imipenem (16.7%) was seen amongst Klebsiella isolates recovered from tracheal aspirate. Of the 100 Klebsiella isolates, 41 (41%) demonstrated resistance or reduced susceptibility to ceftazidime and/or aztreonam, of which 35 isolates were identified as ESBL-producer Klebsiella strains by the double-disk synergy test; 26/75 were recovered from urine (34.6%), 7/12 from tracheal aspirate (58.3%) and 2/7 from sputum (28.6%).
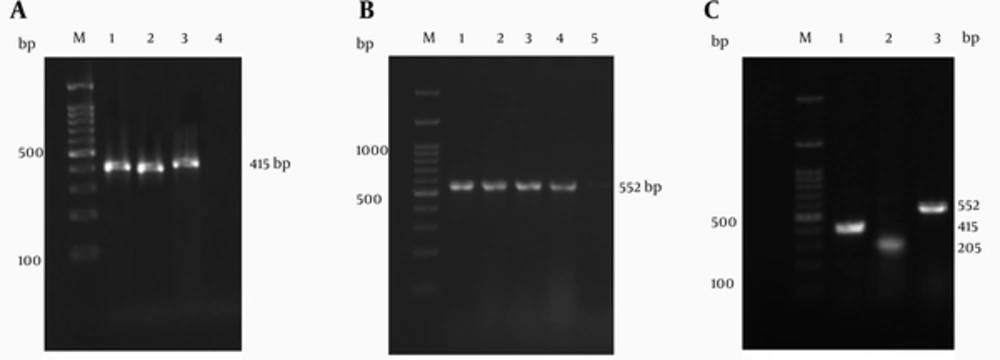
Twenty-eight (80%) of the ESBL-producing Klebsiella isolates were found to carry blaCTX-M type genes. According to the PCR assay and sequencing, 21 (60%), 15 (42%) and nine (34%) of these CTX-M positive Klebsiella isolates, were identified as CTX-M-1, CTX-M-2 and CTX-M-9 types, respectively (Figure 1). Also none of the isolates were positive for CTX-M type 8. The majority of CTX-M positive Klebsiella isolates were CTX-M-1. Eight (22.9%) of the CTX-M positive Klebsiella isolates carried both blaCTX-M-1 and blaCTX-M-2 genes and three (8.6%) were positive for blaCTX-M-1, blaCTX-M-2 and blaCTX-M-9 genes.
Lanes M: 100-bp DNA ladder as the molecular size marker; (A) Lane 1: positive control; Lanes 2 and 3: blaCTX-M-1 positive Klebsiella isolates; Lane 4: negative control; (B) Lane 1: positive control; Lanes 2 to 4: blaCTX-M-2 positive Klebsiella isolates; Lane 5: negative control; (C) Lane 1: blaCTX-M-1 positive Klebsiella isolate; Lane: 2: blaCTX-M-9 positive Klebsiella isolate; Lane 3: blaCTX-M-2 positive Klebsiella isolate.
The resistance of blaCTX-M-1 carrying Klebsiella isolates to antibiotics was 100% to ampicillin, 80% to aztronam, 79% to amoxicillin-clavulanic acid, 76% to cotrimoxazole, 71% to ceftazidime, 71% to cefoxitin, 65% to ceftriaxone, 25% to ciprofloxacin and 1.2% to imipenem. GenBank accession numbers obtained for the purified PCR products are KJ803828 to KJ803829, respectively.
5. Discussion
The number of CTX-M-ESBL-types has rapidly increased worldwide. The incidence of CTX-M-ESBL-producing bacteria has also rapidly increased worldwide (13). Klebsiella spp. has been reported as the most prevalent ESBL-producing bacteria (14, 15). Different studies have shown that ESBL-producing Klebsiella are commonly isolated from urine and blood (16). In the present study, ESBL production was most frequently seen amongst Klebsiella strains isolated from tracheal aspirate. In a study conducted during year 2009 in Tehran, all K. pneumoniae isolated from blood or eye specimens were ESBL producers (17). According to current documents any bacterium which is confirmed as an ESBL producer, should be reported resistant to all extended-spectrum β-lactam antibiotics and carbapenems, which are drugs of choice for the treatment of infections caused by these isolates (18). In our study, 34.6% and 58.3% of Klebsiella isolates, recovered from urine and tracheal aspirate, were identified as multidrug resistant ESBL-producing Klebsiella isolates. Since these isolates showed high resistance rates to drugs of choice such as imipenem, carefully managed therapeutic strategies are required to control Klebsiella spp. urinary and respiratory tract infections.
In the recent years, CTX-M-type ESBLs, were documented as the most dominant-ESBL type worldwide, and have displaced other ESBL enzymes in Enterobacteriaceae (7, 13). We found that 80% of ESBL-producing Klebsiella isolates were CTX-M-type ESBLs. Goudarzi et al. (19) studied 135 E. coli isolates, which were collected from patients with urinary tract infections and showed that 55.5% of the isolates were ESBL producers, and CTX-M-encoding genes were detected in the majority of these isolates. Celenza et al. (20) reported that, among clinical isolates of Enterobacteriaceae in Bolivia, 92% of ESBL-producers were CTX-M-type ESBL producers. The CTX-M-type ESBLs were also more prevalent than any other ESBL types in different studies (21, 22). Since the use of cefotaxime and ceftriaxone is prevalent worldwide, this finding is logical. The PCR and sequencing analysis revealed that the majority (60%) of CTX-M β-lactamases were the CTX-M-1 type. In a study conducted by Edelstein et al. (12) at Russian hospitals and in accordance with our findings, 92.9% of CTX-M β-lactamases were found to be the CTX-M-1 type. In contrast, CTX-M-1 type was reported as the less prevalent subtype of blaCTX-M in ESBL-producing K. pneumonia in China (21). Najar Peerayeh et al. (8) reported that 63.5% of ESBL producer E. coli isolates carried blaCTX-M-1. This subgroup of blaCTX-M was prevalent in our country (14, 23). The high prevalence of CTX-M-1 enzyme amongst the Enterobacteriaceae family in our region could be due to clonal spread of a single clone or patient-to-patient transmission.
Our findings showed that the second most common CTX-M type in ESBL-producing Klebsiella isolates in our region was the CTX-M-2 type, which is in agreement with the findings of Safari et al. (23) conducted during 2012 in the Markazi province. The CTX-M-2 type enzyme has been reported as the most abundant CTX-M type variant amongst Enterobacteriaceae in Latin America (20), and the predominant ESBL type from Argentina (22). The CTX-M-9 type was identified in 34% of our CTX-M-producing Klebsiella isolates. Currently, the CTX-M-9 has been documented as one of the most widespread CTX-M enzymes (7, 24). In Spain and East Asia, the CTX-M-9 enzyme was reported as one of the most prevalent ESBLs (25-27). The association of this CTX-M gene group, with mobile genetic elements such as plasmids and transposons could be the reason. Nevertheless, more studies such as molecular clonality assessment will be required for better understanding of the current dissemination of epidemiologically important CTX-Ms such as CTX-M-9. The CTX-M-8 type was not detected in our ESBL-positive Klebsiella strains. Similar to the present study, the CTX-M-8 related enzymes were not found amongst nosocomial isolates of E. coli and K. pneumoniae in the study conducted by Edelstein et al. (12) yet in contrast to our findings, the presence of CTX-M-8 among ESBL positive isolates of Enterobacteriaceae was reported as 17.5% in Markazi province (23).
CTX-M-type ESBLs, of different genetic groups, have been reported by other studies from Spain, France, Japan and Korea, showing that the pattern of CTX-M-type ESBL genes could vary geographically (27-30). The results obtained by the PCR method showed that eight CTX-M-positive Klebsiella isolates harbored both blaCTX-M-1 and blaCTX-M-2 genes, and blaCTX-M-1, blaCTX-M-2 and blaCTX-M-9 genes coexisted in three CTX-M positive Klebsiella isolates. The associations of several β-lactamases have been documented in multi-drug resistant K. pneumoniae isolates (21). Amongst blaCTX-M-1 carrying Klebsiella isolates, a high level of resistance was seen to ampicilin, aztronam, amoxicillin-clavulanic acid, cotrimoxazole, ceftazidime, cefoxitin, ceftriaxone and ciprofloxacin. As the CTX-M-1 type was reported as the most prevalent subtype of CTX-M-ESBLs amongst ESBL-producing Klebsiella isolates in our region, this finding provides useful information for the treatment of infections caused by Klebsiella strains.
In conclusion, our study revealed that the frequency of blaCTX-M genes among Klebsiella isolates in our region was at an alarming rate showing that epidemiological monitoring is necessary. Also the majority of ESBL genotypes in our Klebsiella isolates was blaCTX-M-1.