1. Background
Nowadays, hepatitis C virus (HCV) infection represents a major public health concern, affecting over 170 million individuals worldwide (1, 2). Infected patients may be at risk of progressive liver fibrosis and cirrhosis and hepatocellular carcinoma (HCC), as well as life-threatening and irreversible extrahepatic injuries, even after transplantation and treatment (3-5).
Although novel therapeutic regimens to combat HCV infection have been developed which can cure a large proportion of chornically infected patients (2), important concerns related to HCV infection control still exist. First, therapy in infected patients with HCV genotype 3, known as the second most prevalent and the most-difficult-to-treat type, resulted in the lower rate of sustained virological response (SVR), compared with other genotypes (6, 7). Secondly, the poor compliance to the treatment protocols due the high cost and relatively long period of treatment may result in the most patients’ refusal to accurately follow up or even accept the treatment protocols (8, 9). Therefore, it is estimated that the infected population will increase further in the next few years (10). Consequently, using treatment as an effective control strategy could not completely interrupt the transmission and decrease the disease burden; so, more attempts to develop other low-cost but efficient HCV preventive strategies such as vaccines are needed.
Hepatitis C virus belongs to the Flaviviridae family with a positive-sense RNA genome which encodes different structural and non-structural proteins (11). It was shown that high levels of viral genome mutation lead to heterogeneity (12), as well as some modifications in virus regulatory elements (13). Moreover, the creation of new subtypes among different genotypes of the virus is also highly possible (14). Therefore, the development of HCV universal vaccine is faced with major challenges and no vaccine still exists (15).
To date, DNA vaccines as the safest and most promising means are designed or under clinical trials to elicit host immune responses (humoral and cellular) against HCV, as well as HIV and Influenza (16). Previous study confirmed HCV-specific immunogenicity following vaccination with a DNA vaccine candidate harboring immunodominant Core, E2, NS3 and NS5B HCV epitopes in BALB/c mice (17). It was revealed that the antigenic epitopes of the target proteins expressed by DNA vaccine plasmids more closely resembled the native viral proteins than those of traditional vaccines such as the attenuated and subunit ones (16). Hepatitis C virus DNA vaccination has been useful for prevention or even as a therapeutic way to control such infections by activating T-helper and cytotoxic T cells, as well as antibody responses in animal models (16), but genotype 1 of the virus has been more studied. Limited research has been carried out on designing DNA vaccines for genotype 3.
2. Objectives
Paving way to develop a novel DNA vaccine candidate for HCV genotype 3a, the current study aimed to construct a eukaryotic expression vector encoding NS3/NS4A nonstructural proteins of the respective genotype and evaluate its expression in Huh7 cell line.
3. Materials and Methods
3.1. Construction and Identification of Recombinant Plasmid
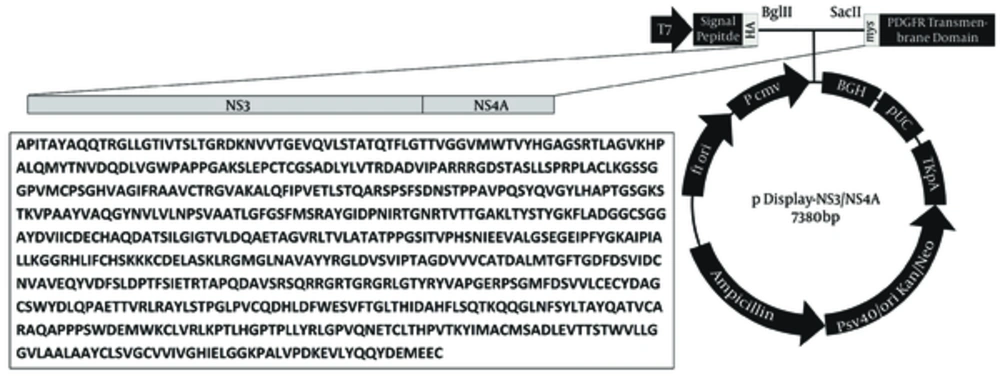
A set of primers were designed according to the 14 available NS3/NS4A nucleotide sequence data of 3a subtype of HCV from the GenBank database of the National Center for Biotechnology Information (NCBI). The sequences were initially analyzed by Lasergene sequence analysis software package (DNAStar, Madison, WI, USA); the consensus sequence for NS3/NS4A was generated using Clustal X (version 1.8) software and the primer set was designed based on the result (forward NS3/4A: 5’-AGATCTGCCCCGATCACAGCATACGCCC-3’; reverse NS3/4A: 5’-CCGCGGGCACTCCTCCATCTCATCG -3’ caring, respectively, the BglII and SacII cloning sites (underlined and boldface)). Viral RNA was extracted using commercially available kit (Invitek, Berlin, Germany) from 200 µL plasma of a patient infected with HCV genotype 3a, confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR) and nested PCR, according to the method of Ohno et al. (18). The extracted RNA was used for cDNA synthesis using cloned avian myeloblastosis virus (AMV) reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and PCR was performed using Platinum® Taq DNA polymerase high fidelity (Invitrogen, Carlsbad, CA, USA) in a 25 μL reaction. The amplified NS3/NS4A fragment was cloned into pTZ57R/T intermediate cloning vector (Fermentas, Lithuania) and transformed into Escherichia coli DH5α competent cells (TaKaRa Biotechnology Co., Dalian, China).
The respective recombinant plasmid was extracted and cleaved by BglII and SacII (Fermentas, Lithuania) and inserted into the similarly digested eukaryotic expression vector pDisplay (Invitrogen, Carlsbad, CA, USA) with T4 DNA ligase (Invitrogen, Carlsbad, CA, USA) and transformed into E. coli DH5α. The pDisplay vector contains hemagglutinin A (HA) epitope tag in upstream and myc epitope in downstream of the cut sites which allow for the detection of the expressed recombinant proteins by immunofluorescence assay using anti-HA/myc antibodies. PCR and restriction endonuclease assays and sequencing in ABI 3130 Genetic Analyzer by BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) were used to screen, identify, and confirm positive clones in both cloning steps. The resulted recombinant plasmid was named pDisplay-NS3/NS4A.
3.2. Cell Transfection
The Huh7 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Germany) containing 10% fetal bovine serum (Gibco, Germany) at 37°C and 5% CO2. The logarithmic phase of the cells was harvested and seeded into six-well culture plates (2 × 105 cells/well) with 2 mL of the plating medium to become 70% - 80% confluent. The cells were transiently transfected with 4.0 µg purified pDisplay-NS3/NS4A plasmid, using 10 µL Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
3.3. Immunocytochemical Staining
After 48 hours of transfection, the cells were permeabilized and the expression of the proteins was determined using mouse monoclonal antibody to the C-terminal HA tag and goat polyclonal antibody to N-terminal myc tag of the fusion construct, according to the manufacturer’s guideline (Abcam, UK). The expression of the fusion proteins were evaluated under fluorescence microscope. The nontransfected cells were considered as blank, while empty pDisplay-transfected cells were considered as negative control (mock) and both were stained with anti-HA and -myc FITC-conjugated antibodies. The sequences of the HA/myc tags in the empty pDisplay vector are not in-frame for expression, distinctly. They can only be expressed if their genes are completed with the in-frame inserted sequence containing the sticky ends of the recommended restriction enzymes. Membrane fluorescence was detected using Zeiss Axioskop 2 fluorescence microscope (Zeiss, Göttingen, Germany).
4. Results
4.1. Evaluation of the Reverse-Transcriptase Polymerase Chain Reaction Product and Recombinant pDisplay-NS3/NS4A Eukaryotic Expression Vector
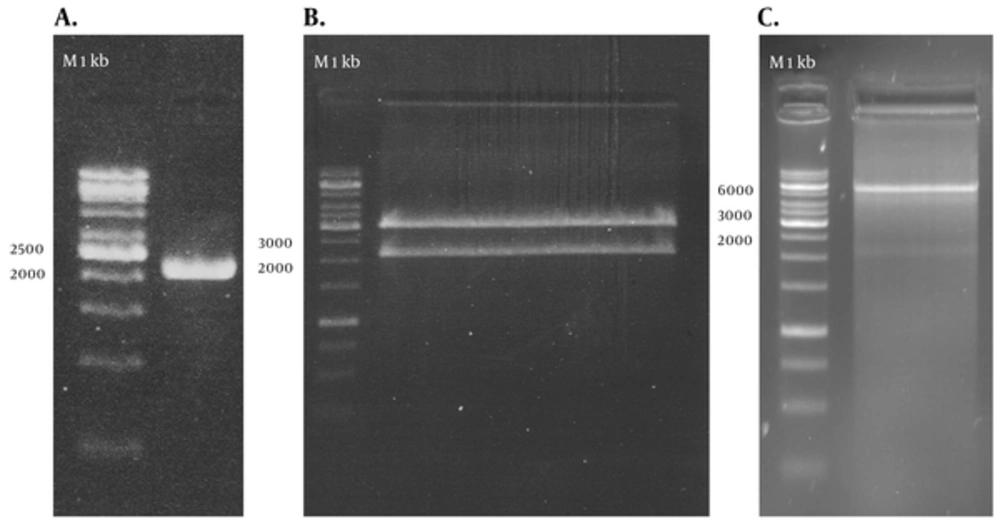
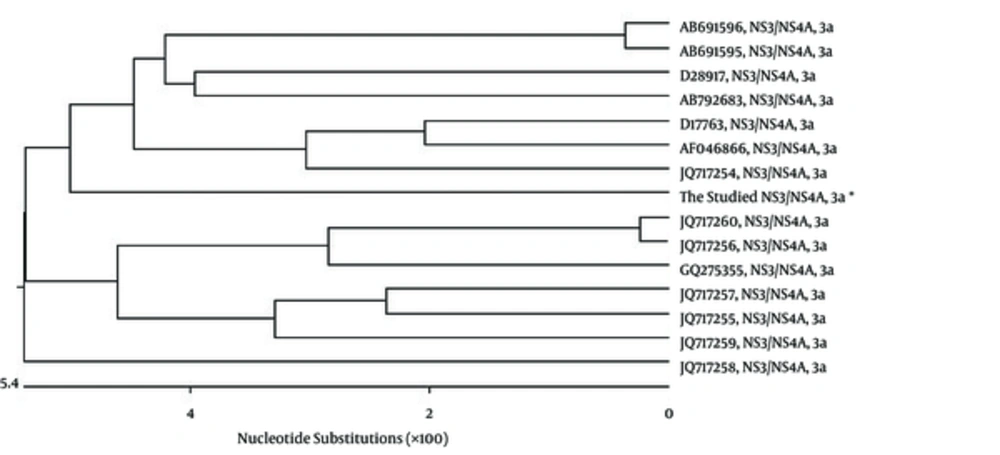
RT-PCR amplified the NS3/NS4A fragment and the products were loaded on 1.5% agarose gels; the band for full-length of the respective cDNA was 2055 bp (Figure 1 A). The fragment was successfully cloned into the intermediate cloning vector (Figure 1 B) and then into pDisplay eukaryotic expression vector (5325 bp, Figure 1 C). The fragment was confirmed by BglII and SacII digestion and electrophoresis. Besides, the obtained DNA sequencing results confirmed the cloned gene completion, correct position (Figure 2), and the analysis revealed its high homology with the sequences of NS3/NS4A of HCV genotype 3a database in GenBank (Figure 3).
A: PCR amplification of NS3/NS4A using specific primers, 1 kb DNA ladder (Vivantis, Malaysia). B: recombinant T/A cloning vector digested by BglII and SacII enzymes, showing the 2055 bp insert, 1 kb DNA ladder (Fermentas, Lithuania). C: pDisplay-NS3/NS4A plasmid digested by BglII and SacII enzymes, showing the 2055 bp insert, 1 kb DNA ladder (Fermentas, Lithuania).
4.2. Evaluation of Protein Expression of pDisplay-NS3/NS4A in Huh7 Cells
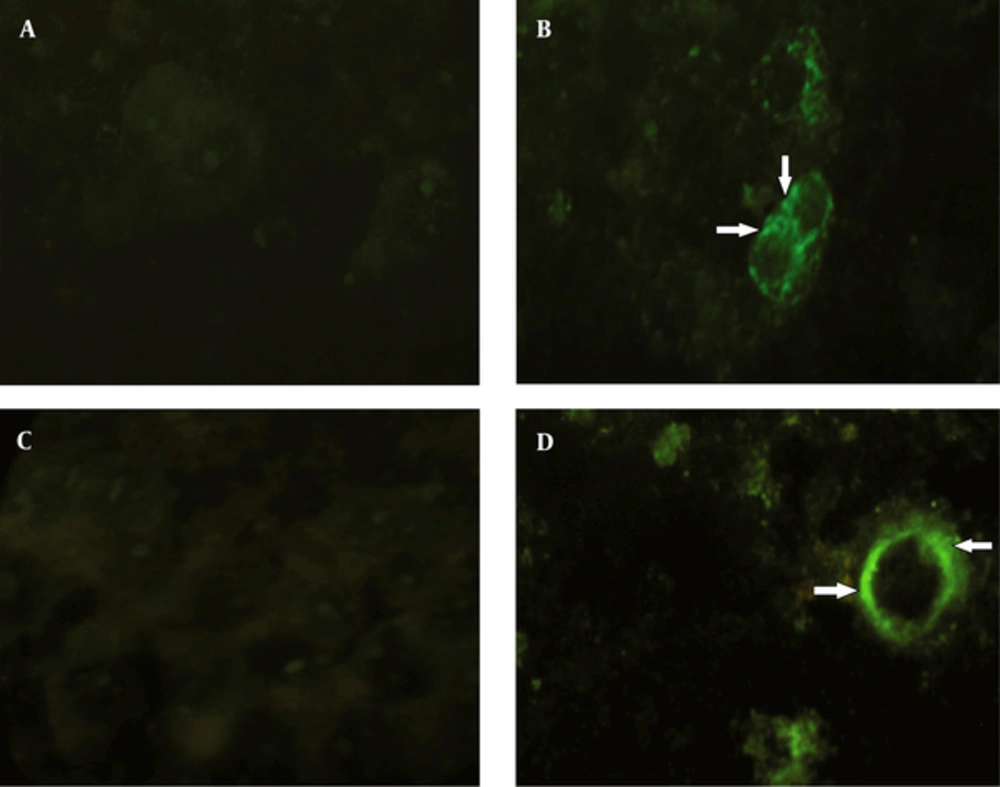
After transfection, the expression of pDisplay-NS3/NS4A was analyzed by immunocytochemical staining. The results of immunofluorescent assay using antibodies to hemagglutinin A tag indicated that the respective protein was being successfully expressed on the surface of the Huh7 cells, when compared with the control group. The result of the assay using antibodies to myc tag was the same (Figure 4).
A: no immunofluorescence was detected in mock-infected Huh7 cells using conjugated antibodies to the HA tag. B: HA expression and its specific localization (arrows) at the cell surface membrane of the Huh7 cells. C: no immunofluorescence was detected in mock-infected Huh7 cells using conjugated antibodies to the myc tag. D: myc expression and its specific localization (arrows) at the cell surface membrane of the Huh7 cells.
5. Discussion
Recently, the utilization of the nonstructural genes as a DNA vaccine has revealed quite promising results in lab animal models (19) and also in clinical trials (20). Previous studies demonstrated that the resolution of acute infection was closely associated with the specific T cell responses to different HCV proteins (21, 22). The comparison of the peripheral blood mononuclear cell responses to the recombinant HCV proteins core, NS3, NS4, and NS5 among the patients who spontaneously clear the virus showed that the correlation was remarkably high with responses to NS3 (22). Although this protein is able to down-regulate the innate immune responses by various pathways such as decreasing the expression of interferon stimulatory genes (ISGs) (23), owing to its multiple important epitopes with the ability to induce potent immune responses, it has been recognized as one of the central targets of interest for new vaccine developments (17, 24). A study by Lang et al. in both mice and Rhesus macaques demostrated strong HCV NS3-and NS4A-specific cellular immune responses preceeded by the injection of DNA vaccine encoding the respective proteins (19). Therefore, it seems that nonstructural proteins play an important role in developing possible future HCV DNA vaccines. In the present study, the NS3/NS4A fragment of the nonstructural genome of the virus was applied in the construction of our eukaryotic expression vector.
Most of the previous HCV DNA vaccine studies were mainly focused on genotype 1 of the virus (25, 26) and to our knowledge limited research has been performed on genotype 3. A recent study on the global determination of HCV major genotypes distribution reported that HCV genotype 1 was the most prevalent worldwide (46.2%), followed by genotype 3 (30.1%) (27). Similarly, in Iran, it was shown that genotypes 1 and then 3 were the most prevalent HCV genotypes among both nontransplant (28) and transplant patients (29). Hence, in the present study, the nonstructural proteins of genotype 3a of the virus were investigated. The reason of utilizing NS4A protein as a fusion peptide combined with the NS3 protein in our study was its reported roles in the enhancement of stability, expression, and immunogenicity of the NS3 protein. It was revealed that in the absence of NS4A, the full-length NS3 protein will be diffusely distributed in the cytoplasm and nucleus and rapidly degraded (30). The very hydrophobic amino-terminal domain of NS4A assists in the NS3-NS4A complex anchoring and localization in the membrane and may protect it against proteolytic attack (31). Therefore, co-expression of NS4A will improve the survival of NS3 and its stability in cellular context, and consequently the immunogenicity of the respective protein (32). Although the rationale behind the observation in increasing of NS3 expression in the presence of NS4A was not determined (32); it may be related to lower degradation rate of the NS3 protein after co-expression with NS4A.
Selection and designing the DNA vaccine vectors play an important role in increasing the efficiency of vaccination (33). Numerous kinds of eukaryotic expression vectors have been widely applied in this system. We used pDisplay as a eukaryotic expression vector in this study, due to some of its documented benefits including: (i) the vector is under the control of the human cytomegalovirus (CMV) immediate-early promoter/enhancer which helps to increase the expression level of the recombinant protein (34); (ii) the HA and myc epitopes on both sides of the expressed recombinant proteins allow the detection of the fusion peptide with anti-HA/myc antibodies; (iii) the murine Ig k chain leader sequence and platelet-derived growth factor receptor transmembrane domain (PDGFR-TM), located at the N-terminus and the C-terminus, respectively, which direct the fusion protein to the secretory pathway and anchor it to the plasma membrane (35). In the present study, the results of the immunofluorescent assay confirmed that the target protein anchored to the Huh7 cell membrane. Previous studies on immunization against other pathogens with DNA vaccines revealed that the antigenic peptide produced on the cell surface was thought to be more immunogenic than those secreted or expressed as intracellular forms (36, 37). This may be resulted from the higher chance of antigen capturing and processing by antigen presenting cells (38). In most of the earlier studies on HCV DNA vaccines, the target proteins were expressed within the cell, not on the cell surface (39, 40).
Considering the future aim to test and develop a DNA vaccine candidate against HCV genotype 3a, we designed, constructed and evaluated the in vitro expression of a eukaryotic expression vector, coding NS3/NS4A proteins of the virus. In this fundamental experiment, the target fragment of HCV genotype 3a was successfully reverse transcribed, amplified, cloned into a eukaryotic expression vector, and expressed in Huh7 mammalian cells. Further research is underway to evaluate this fragment immunogenicity in lab animal models.