1. Background
Respiratory syncytial virus (RSV) is an enveloped, single-stranded, negative-sense RNA virus with a genome of ∼15,200 bases and a member of the genus Pneumovirus in the Paramyxoviridae family. Based on antigenic and genetic variability, RSV is classified into two groups, A and B (1, 2). Respiratory Syncytial Virus is the major cause of lower respiratory tract infections (LRTIs) with considerable morbidity and mortality in young children, the elderly, immunocompromised individuals, and patients with chronic lung diseases (1, 3, 4). There is some evidence supporting the theory that early LRTI with RSV raises the chance of developing chronic asthma-like respiratory symptoms during childhood (5). With this high disease burden and the lack of an effective RSV treatment and vaccine, there is a clear need for discovery and development of novel, effective and safe drugs to prevent and treat RSV diseases (4, 6, 7).
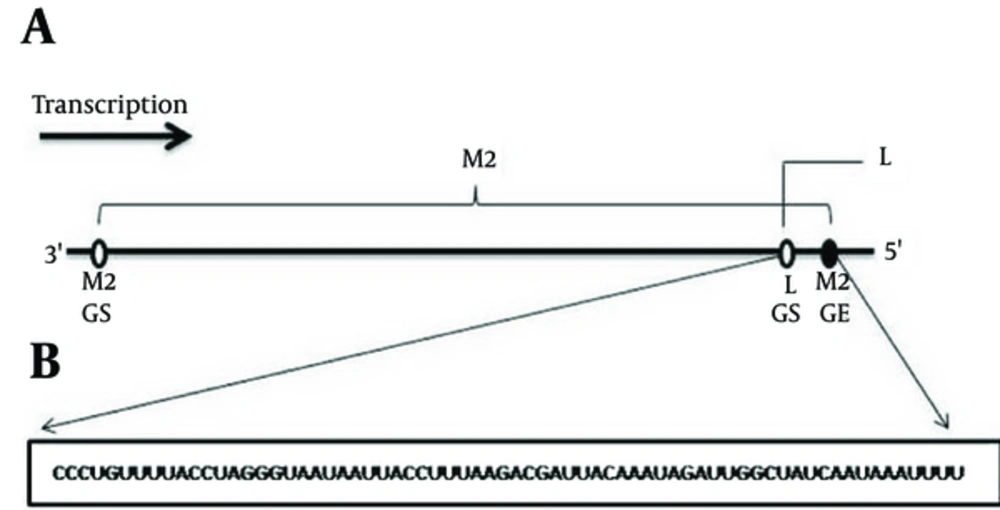
The RSV gene order is 3’-NS1-NS2-N-P-M-SH-G-F-M2-L-5’ (8). There are several conserved motifs at the boundaries of each gene, including the gene start and gene end signals which direct transcription initiation and termination, respectively (9). Viral proteins are produced by RSV-specific, RNA-dependent RNA polymerase (RdRp) that is packaged into the nucleocapsid (10). The nucleocapsid is the necessary unit for RNA replication and production of progeny virus particles (5). Respiratory syncytial virus protein sequence diversity studies showed that the surface protein G is highly divergent between strains and the internal proteins, such as neucleoprotein (N), are highly conserved, with more than 90% homology (5, 11). One distinctive feature of RSV is the 68 nucleotide overlap in the M2 and L genes, so that the M2 gene-end (GE) signal lies downstream of the L gene-start (GS) signal (9). The invention of RNA interference (RNAi), an endogenous and ubiquitous pathway, leads to revolutionary new concepts in human therapeutics (12). Since the discovery of RNAi, there has been rapid progress toward its use as a therapeutic approach against human diseases (13, 14).
There are two separate pathways of RNAi mechanisms, the siRNA (small interfering RNA) and the microRNA which can be used to effectively suppress the expression of genes in a sequence specific manner (15). The siRNA pathway, which exploits only cytoplasmic processes, is now the primary pathway utilized in RNAi-based drug development (12). siRNA-based drugs have distinct advantages over conventional small molecule or protein-based drugs, including high specificity, higher potency, and reduced toxicity (16, 17). Several key criteria are considered for selection of potent and functional siRNA molecules, such as the GC content of the sequence, specific nucleotides in distinct positions, sequence conservation, thermodynamic parameters of siRNA duplexes, and circumventing possible off-target effects (18).
Several studies have shown that siRNA can function as miRNA and regulate unintended transcripts. Such off-target effects via seed complementarities in their 3’UTRs can produce false positives in siRNA screens. In this regard, accurate prediction of miRNA-like off-target effects is important to mitigate undesired results (19, 20). siRNAs are loaded into the RNA-induced silencing complex (RISC) to cleave target mRNAs that share sequence identity with the siRNA. Within argonaute2-directed RISC, the siRNA unwind and antisense (guide) strand directs RISC to target the mRNA sequence after the sense (passenger) strand is released and cleaved by cellular nucleases (21, 22).
2. Objectives
The present study aimed to design potential siRNAs for silencing nucleoprotein and an overlapping region of M2-L coding mRNAs of RSV by computational analysis.
3. Materials and Methods
3.1. Sequence Retrieval and Multiple Sequence Alignment
Two hundred and nineteen complete CDS of N genes of different human RSV strains were collected from the viral gene bank database available at National Centre for Biotechnological Information (http://www.ncbi.nlm.nih.gov/). The accession numbers of these complete CDS are available upon request. The accession number of the sequence used for retrieval of the sixty-eight nt overlapping region of the M2-L gene was AF254574.1. In comparison to the N gene, the M2-L overlapping region is highly conserved in different RSV strains, then just one RSV reference strain was used for siRNA designing. Figure 1 depicts the structure of the M2/L overlap. Using MEGA5, all the complete CDS (219) were aligned for the selection of conserved regions. As one mismatch between the target mRNA and siRNA could dramatically affect siRNA efficiency, this method ensures the design of siRNAs that target all the consensus regions of transcripts. The regions in the target site with the highest homology by the mRNAs are considered in the following analysis.
3.2. Potential siRNA Designing
In order to design siRNAs, two forms of software were used: the Whitehead siRNA selection program (http://sirna.wi.mit.edu/home.php) and Invitrogen BLOCK-iTTMRNAi designer (https://rnaidesigner.lifetechnologies.com/rnaiexpress/). The Whitehead siRNA selection website provides the flexibility of using several common siRNA sequence patterns (e.g. A: AAN19TT, B: NAN19NN, C: N2(CG)N8(AU)(N8(AU)N2) or a custom pattern (23). All of the siRNAs were aligned to find the most common siRNAs. The siRNA design rules can be categorized into both sequence and structural rules. Briefly, sequence rules consider that the target region should be preferably 50 - 100 nt downstream of the start codon because those regions are usually occupied by diverse protein factors (24). The G/C content of the binding site and siRNA should be in the range of 45% - 65% which provides the necessary stability for the siRNA (18). Numerous sequence rules regarding the preference or avoidance of the specific positions of the sense or antisense strand of the duplex have been suggested (24). Structural rules specify the structural accessibility of the target site and the thermodynamic features of the siRNA/target duplex. Apart from using the siRNA design tool, the resulting siRNAs were manually analyzed according to the parameters recommended by Ui-Tei et al. (25), Reynold et al. (26) and Jagla et al. (27) for optimal design.
3.3. Similarity Search
A BLAST search was used to compare a query sequence of our designed siRNAs with human genomic and transcripts database (http://blast.ncbi.nlm.nih.gov/) to identify any off-target sequence similarity in other non-targeted genomes by applying expected threshold value 10 and BLOSUM 62 matrix as parameters. siRNA with 15 or more consecutive bases of homology with any other mRNAs were excluded from consideration.
3.4. GC Calculation and Secondary Structure Prediction
For GC content calculation of predicted siRNA, the Oligo Analyzer Calculator (https://eu.idtdna.com/calc/analyzer) was used. The GC content of the siRNA duplex is a candidate which might correlate with siRNA functionality. Too high GC content may slow down duplex unwinding and too low GC content may decrease the efficiency of target mRNA recognition and hybridization (28). The Mfold web server (http://www.mfold.rna.albany.edu/) was used for secondary structure prediction.
3.5. Calculation of RNA-RNA Interaction Through Thermodynamics
For computing the free energy of siRNA folding and studying the thermodynamics of interaction between the predicted siRNA and the target gene, the RNAcofold program was used. RNAcofold calculates the hybridization energy and base pairing form of two RNA sequences (29).
4. Results and Discussion
RNAi methodology can be selected to target viral genes with high levels of sequence conservation. Nucleoprotein (∼1175 bp) is known as the most conserved gene of RSV that encapsidates the genome for protection from nuclease. The encapsidated genomic RNA serves as a template for transcription and replication (30). In transcription, subgenomic mRNAs were produced by a stop-start mechanism and, in replication, genome-length positive strand antigenomes were produced by read-through synthesis (30, 31). A key feature of both the genome and antigenome templates is that they remain coated with N protein all the time which, aside from preventing the formation of secondary structures in the RNA also protects them from nuclease attack (32). It can be considered that silencing of this gene will abrogate its essential functions.
In the present study, about 219 gene sequences from different strains of RSV were obtained from the GenBank, NCBI. The sequences were subjected to the MEGA5 computational tool in order to find the consensus regions of selected sequences. Nucleoprotein conserved regions were taken into account for optimal siRNA selection. Whitehead siRNA selection software and the Invitrogen BLOCK-iTTMRNAi Designer were used to provide functional siRNA design. Whitehead software also predicts off-target effects based on the seed complementarity of the target site among related species.
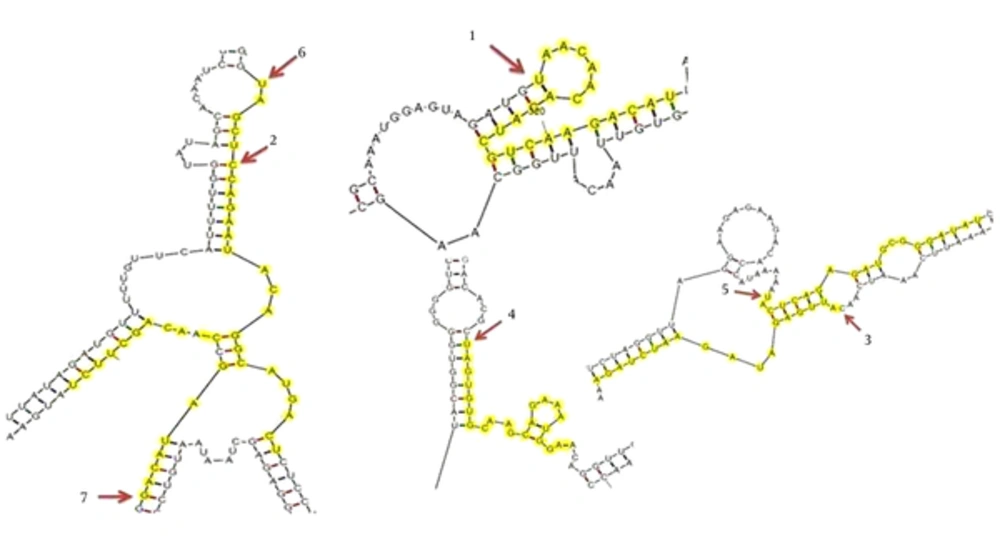
Fifty-six siRNA suggested by the Whitehead server and ten siRNA suggested by Invitrogen were manually analyzed for optimal design (mentioned in part 3.2 in methods). According to this system, a score of six or more significantly increases the probability for gene silencing. Finally, seven siRNAs were found to meet the rules and these are presented in Table 1. The siRNAs were sorted on the basis of targeting the largest number of nucleoprotein genes. All siRNAs were subjected to BLASTN to ensure that the siRNAs were specific for RSV. All the siRNAs retain GC content within 36% - 48%. Too high and too low GC content can impede the loading of siRNAs into RISC and weakening hybridization between the siRNA and target mRNA, respectively (33).
| Target No. | Location of Target Within mRNA | siRNA Target Within mRNA | Predicted siRNA Duplex Candidate | Length and Pattern | GC, % | Tm, °C | Free Energy of Folding With Target | ΔG Binding, k/cal | miRNA Target | No. of Targeted Sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 290 - 311 | UAACAACACAUCGUCAAGACAU | 22 (B, C) | 36.4 | 41.4 | 0.4 | -34.8 | 204 (78) | 150 | |
| UAACAACACAUCGUCAAGACAU | ||||||||||
| CCAGAATACAGGCATGACT | ||||||||||
| 2 | 439 - 457 | CCAGAATACAGGCATGACT | 19 (C) | 47.4 | 51.8 | 0.6 | -33.9 | 142 (63) | 141 | |
| CCAGAAUACAGGCAUGACU | ||||||||||
| AGUCAUGCCUGUAUUCUGG | ||||||||||
| 3 | 378 - 396 | CATTGAGATAGAATCTAGA | 19 (B,C) | 31.6 | 42.4 | 0.6 | -28 | 95 (45) | 126 | |
| CAUUGAGAUAGAAUCUAGA | ||||||||||
| CAUUGAGAUAGAAUCUAGA | ||||||||||
| 4 | 825 - 845 | TAGTGTGCAAGCAGAAATGGAT | 22 (C) | 40.9 | 46.3 | 0.4 | -37.7 | 116 (53) | 60 | |
| UAGUGUGCAAGCAGAAAUGGAU | ||||||||||
| AUCCAUUUCUGCUUGCACACUA | ||||||||||
| 5 | 245 - 264 | TACTCAGAGATGCGGGATAT | 20 (B, C) | 45 | 47.6 | 0.4 | -36 | 272 (87) | 36 | |
| UACUCAGAGAUGCGGGAUAU | ||||||||||
| AUAUCCCGCAUCUCUGAGUA | ||||||||||
| 6 | 434 - 453 | TAGCTCCAGAATACAGGCAT | 20 (C) | 45 | 53 | 0.4 | -36.2 | 142 (63) | 15 | |
| UAGCUCCUGAAUACAGGCAU | ||||||||||
| AUGCCTGUAUUCUGGAGCUA | ||||||||||
| 7 | 610 - 628 | GAC ATAGCC AAC AGC TTCT | 19 (C) | 47.4 | 52.2 | 0.6 | -33.8 | 198 (77) | 15 | |
| GACAUAGCCAACAGCUUCU | ||||||||||
| AGAAGCUGUUGGCUAUGUC |
Since viruses are known to be genetically diverse (34), some researchers have focused more on conserved target sites to design siRNAs. Rosales et al. have used this approach to design siRNAs against NS4B and NS5 of the Dengue virus (35). Likewise Raza et al. predicted siRNA against the conserved region of HA and NA genes of the Influenza A virus (36). Naito et al. addressed the importance of conserved regions of HIV-1 for siRNA targeting (37).
Ideally, designed siRNAs must be complementary to their target sequences. However, several studies have shown that siRNA can bind to the mRNAs in a miRNA-like way through partial complementarity, leading to undesirable side-effects (38, 39). Here, for each siRNA candidate, the number of genes that the 7mer miRNA seed could potentially bind to is displayed (Table 1, miRNA target). For example, 204 (78): 204 means the miRNA seed (7mer) could potentially bind to 204 genes; 78 means that 78% of all 7mers would potentially bind to those 204 genes. In other words, only 22% of all 7mers would bind to more than 204 genes. This can be very useful for explaining the off-target effects in the siRNA experimental results.
siRNA sequences that contain internal repeats or palindromes may reduce the effective silencing by forming fold-back structures (40). Relative stability and the propensity to form internal hairpins can be predicted by melting temperature (Tm). Sequences with high Tm are prone to form hairpin structures. So, duplexes lacking stable internal repeats are better silencers (Tm < 60°C) (26).
RNA structure prediction of target mRNA is an important tool for designing siRNA. The accessibility of the target site can be important for estimating the effectiveness of the small RNA for regulating gene expression (41). A linear correlation has been reported between increasing stability of secondary structures and decreasing cleavage efficiency. Indeed, secondary structures which limit the accessibility of the target site by direct blockage reduce RISC-mediated cleavage efficiency (42). Using Mfold software, the secondary structure of nucleoprotein was predicted and the interaction sites with the proposed siRNAs are shown in Figure 2. It can be concluded that siRNA numbers 1, 3, 5 and 6 have more accessibility for target recognition.
RNAcofold computes the hybridization energy and base pairing pattern of interacting RNA molecules by a combination of thermodynamic and kinetic considerations. It provides an extension of McCaskill’s partition function algorithm to compute base pairing probabilities (29). So, for the interaction of a target mRNA and its predicted siRNA, free energy of binding is calculated. The target-siRNA complex product prefers negative overall ΔG. A stable duplex has more negative ΔG than an unstable one (Tables 1 and 2).
| Target No. | Location of Target Within mRNA | siRNA Target Within mRNA | Predicted siRNA Duplex Candidate | Length and Pattern | GC, % | Tm, °C | Free Energy of Folding With Target | ΔG binding (k/cal) | miRNA Target |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 - 32 | GATCCCATTATTAATGGAA | 20 | 32 (C) | 53 | 0.4 | -28.7 | 263 (86) | |
| GAUCCCAUUAUUAAUGGAA | |||||||||
| UUCCAUUAAUAAUGGGAUC | |||||||||
| 2 | 9 - 31 | GGATCCCATTATTAATGGA | 22 | 37 (C) | 46.3 | 0.6 | -30 | 182 (74) | |
| GGAUCCCAUUAUUAAUGGA | |||||||||
| UCCAUUAAUAAUGGGAUCC | |||||||||
| 3 | 35 - 57 | CTAATGTTTATCTAACCGA | 20 | 32 (C) | 47.6 | 0.5 | -26.6 | 1 (1) | |
| CUAAUGUUUAUCUAACCGA | |||||||||
| UCGGUUAGAUAAACAUUAG |
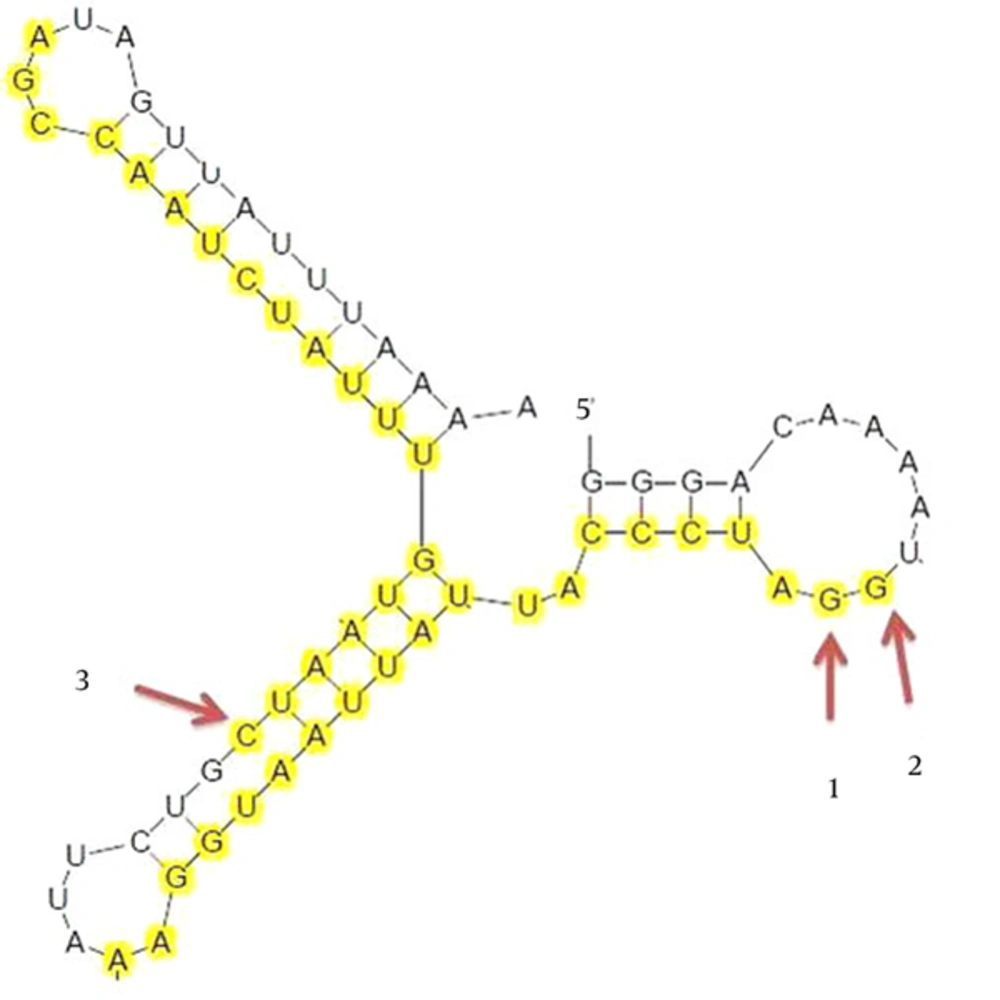
It is worth noting that targeting the overlapping regions of the transcripts simultaneously reduced the level of both transcripts (43). The overlapping region of M2/L mRNA encompasses 68 nt between M2 GE signal and L GS signal. We hypothesized that targeting these two proteins at the same time would likely abolish their function. The properties of the three designed siRNAs targeting the overlapping region of the M2/L gene are summarized in Table 2. A computer-assisted analysis of the secondary structure of the M2/L RNA sequence was performed by Mfold (Figure 3). The results showed that siRNA 1 and 2 have better accessibility to target mRNA.
In conclusion, there is a major unmet medical need for an effective therapy of RSV infection and respiratory viruses may be advantageous to siRNA therapeutics. This paper focused on antivirals that utilize the siRNA pathway as a forefront of drug development. The possibility of turning off the pathogenic genes appears to be an appealing approach to slow down or stop the disease progress of a wide variety of clinical pathogens.