1. Background
Helicobacter pylori is a spiral-shaped, Gram-negative, microaerophilic and fastidious bacterium (1, 2), and infects almost 50% of the world’s population (3, 4). It is the main cause of chronic gastritis, gastric and duodenal ulcers, mucosa-associated lymphoid tissue lymphoma and gastric adenocarcinoma (2, 5). In the international agency for research on cancer (IARC), H. pylori has introduced as a first class (definitive) carcinogen (6). There are several methods for the detection of H. pylori in clinical and environmental samples including culture, polymerase chain reaction (PCR), real-time PCR, histology, rapid urease test, serology, stool antigen test, and urea breath test that classified into two categories called invasive and non-invasive tests on the basis of the using endoscopy. On the basis of Maastricht consensus report (Florence IV), endoscopy and biopsy examination should be carried out in older patients and in patients with alarming sign and symptoms like weight loss, dysphagia, GI bleeding, abdominal mass and iron deficient anemia (7, 8).
Culture, as the gold standard method for diagnosis of H. pylori can diagnose the bacterium to the genus and species level and cultivated isolates can be subjected to antibiotic susceptibility tests. However, this method is not sensitive (70% - 86%), expensive, technically challenging and time-consuming (2, 4). Polymerase chain reaction is also used to detect the H. pylori in clinical samples, but the procedure is complicated and requires expensive instrument, thermal cycler (9). The disadvantages of the rapid urease test include low sensitivity and high amount of false positive results due to production of urea by non H. pylori species (10, 11). Furthermore, in histopathology examination, insufficient number of biopsy specimens and lack of access to samples from different parts of the stomach, the necessity for conducting different staining techniques and time-consuming process may limit the application of this method (12).
Molecular methods for detection of H. pylori from biopsy specimens have been used in research settings but are not applied in clinical practice. Polymerase chain reaction, as a method for detection of the bacterium from biopsy specimens, is being used in many studies (13-15). This method is expensive and time-consuming. Furthermore, the sensitivity of PCR is reduced by very small amounts of contaminating DNA (from a different sample) and inhibitors in the DNA extracted from the samples (1). Loop-mediated isothermal amplification (LAMP) as a new technique for specific amplification of nucleic acid has been described by Notomi et al. in 2000 (16). Loop-mediated isothermal amplification overcomes some drawbacks and limitations of PCR and has been applicable widely in diagnosis test of infectious agents. The method is very specific due to the use of six primers that identify eight regions of the target sequence. The sensitivity of the method for detection of target sequences is 10 times more than PCR. The reaction time of the LAMP is shorter than PCR because LAMP is carried out isothermally (60 - 65°C) and the amplified product is observable without the need to electrophoresis. In addition, LAMP is less expensive than other molecular diagnostic methods because it does not require electrophoresis and thermal cycler. Here, positive result is shown by white insoluble magnesium pyrophosphate and can be seen with the naked eyes (17-19).
2. Objectives
The aim of this study was to set up a LAMP test for detection of H. pylori using designed primers targeted a highly conserved region of the ureC gene.
3. Patients and Methods
3.1. Culture of Helicobacter pylori
In order to setting up the LAMP and PCR reactions, the DNA of a clinical isolate of H. pylori was used. Helicobacter pylori was cultured on enriched egg yolk Columbia agar containing vancomycin (10 mg/L), trimethoprim (5 mg/L) and amphotericin B (5 mg/L). The plates were incubated in microaerophilic condition and 100% humidity at 37°C for 3 to 5 days. Biochemical testes such as catalase, oxidase, urease test and gram staining were used for identification and confirmation of the presumptive colonies (1, 20).
3.2. DNA Extraction
The H. pylori DNA was extracted using high pure PCR template preparation kit (Roche, Germany) according to manufacturer’s instruction. Purity and concentration of the extracted DNA were measured using Nanodrop (Thermo scientific, USA).
3.3. Polymerase Chain Reaction
Species specific primers were designed according to the conserved region of the H. pylori 26695 ureC (glmM) gene as a template (accession No. AE000511) (21). Sequence and properties of the forward and reverse primers were Hp-ureCF: 5’CAT CGC CAT CAA AAG CAA AG3’ (605 - 625 positions in 26695 H. pyloriureC gene) and Hp-ureCR: 5’CAG AGT TTA AGG ATC GTG TTA G3’ (798 - 819 positions in 26695 H. pyloriureC gene). The primers have 100% homology with most reference strains of H. pylori in BLAST.
3.4. Loop-Mediated Isothermal Amplification Primers Design
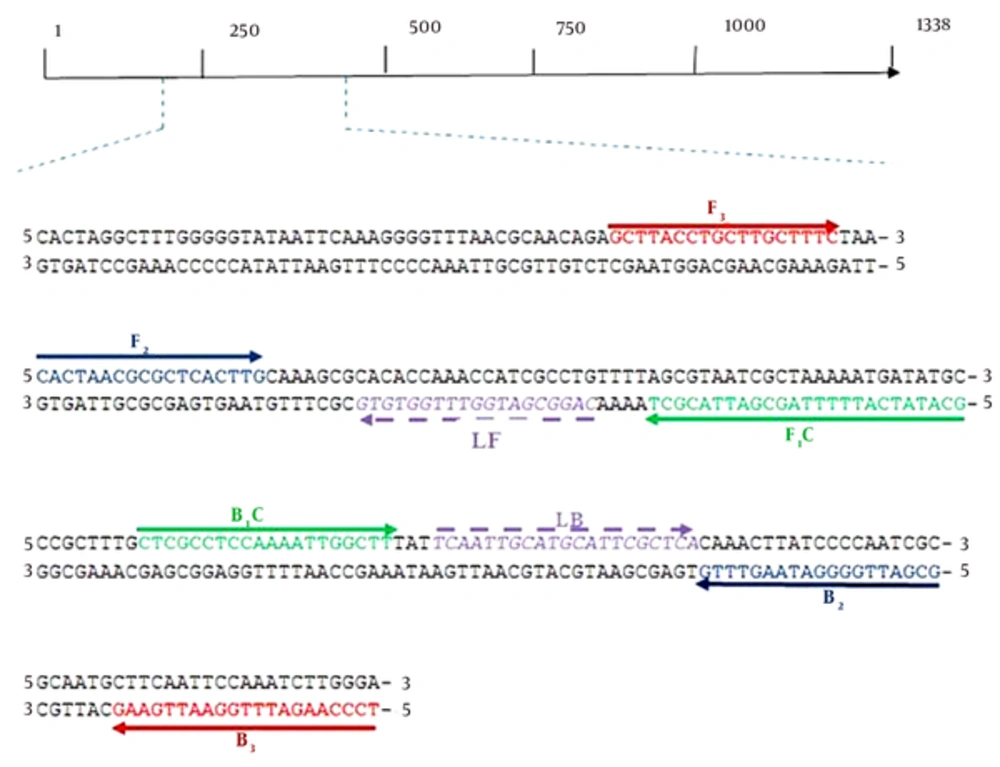
Sequences of the ureC of several strains of H. pylori including 26695 (NC-018939.1), HpaG1 (NC-008086.1), G27 (NC-011333.1) and J99 (NC-000921.1) were retrieved from the GenBank database. In order to obtain the conserved region of the ureC, the retrieved sequences were aligned using Clustalw2 software (www.ebi.ac.uk). Afterward, the primers were designed using the primer explorer software version 4 (www.primerexplorer.jp) based on the conserved sequences (Table 1, Figure 1).
| Primer Name | Sequence (5’ - 3’) |
|---|---|
| ureCFIP | 5’GCA TAT CAT TTT TAG CGA TTA CGC TCA CTA ACG CGC TCA CTT G3’ |
| ureCBIP | 5’CTC GCC TCC AAA ATT GGC TTG CGA TTG GGG ATA AGT TTG3’ |
| ureCF3 | 5’GCT TAC CTG CTT GCT TTC3’ |
| ureCB3 | 5’TCC CAA GAT TTG GAA TTG AAG3’ |
| ureCLF | 5’CAG GCG ATG GTT TGG TGT G3’ |
| ureCLB | 5’TCA ATT GCA TGC ATT CGC TCA3’ |
3.5. Loop-Mediated Isothermal Amplification Reaction
According to the protocol presented by Aryan et al. LAMP reaction was carried out in a final volume of 30 µL containing 0.8 M betaine (Sigma-Aldrich), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 6 mM MgSO4, 0.1% Triton X-100, 2 mM dNTP, 1.6 µM each of the FIP and BIP primers, 0.8 µM of LF and LB primers, 0.2 µM of F3 and B3 primers, 8 U Bst DNA polymerase (New England Biolabs, UK) and 3 µL template DNA (22). In order to set up the LAMP procedure, different temperatures including 63°C, 65°C and 67°C at different periods of time 60, 70 and 80 minutes were tested (23).
3.6. Determination of Analytical Specificity of the Polymerase Chain Reaction and Loop-Mediated Isothermal Amplification
Specificity of the designed primers targeted ureC was evaluated by LAMP and PCR methods using less than 100 ng of extracted DNA from H. pylori and some other bacteria that are commonly found in biopsy samples. The PCR products were subjected to agarose gel electrophoresis and stained with ethidium bromide. Any LAMP reactions that had a white turbidity or a marker-like pattern after agarose gel electrophoresis was considered as positive. A PCR reaction was considered positive if a visible band with 214 bp was appeared on gel electrophoresis.
3.7. Determination of Analytical Detection Limit of Loop-Mediated Isothermal Amplification and Polymerase Chain Reaction
In order to determine the limit of detection of LAMP and PCR, first, the concentration of extracted H. pylori genomic DNA was measured three times, and the mean value was considered as the real concentration. Then, a series of 10-fold serial dilutions of DNA were prepared (1 ng, 0.01 ng, 1 pg, 0.01 Pg, 1 fg, 0.1 fg, 0.01 fg and 1 ag). The optimal PCR and LAMP reaction was carried out by one µL of these samples.
Ultimately, the last dilution of the H. pylori DNA which yielded a detectable band or insoluble turbidity was assigned as the limit of detection of the PCR and LAMP, respectively. To calculate the H. pylori copy number, which was detectable in analytical sensitivity testing the average genome size of H. pylori was considered 1.67 × 106 bp. In fact, 1.67 fg of the H. pylori DNA is equal to a single genome.
4. Results
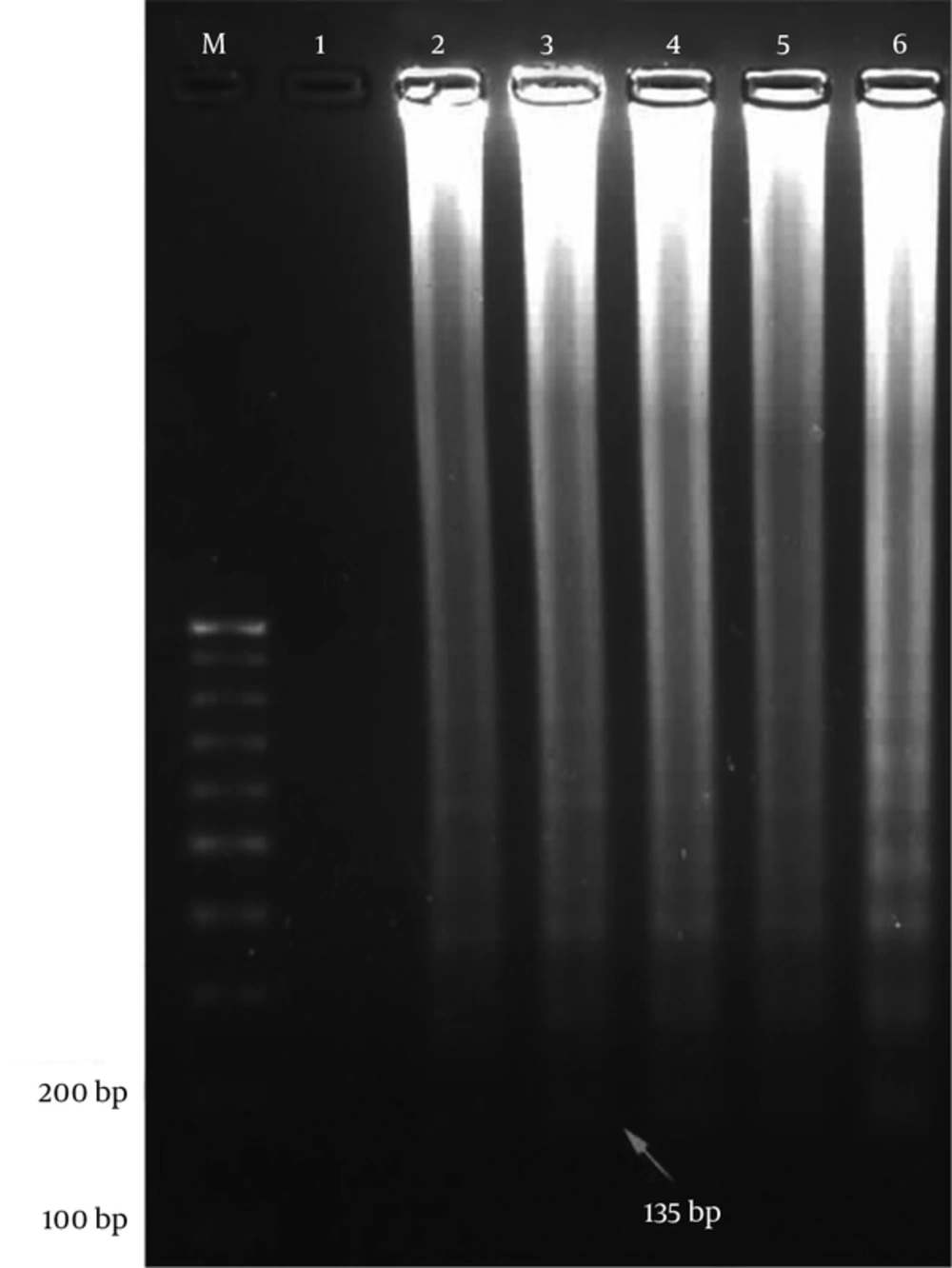
4.1. Identification and Verification of Loop-Mediated Isothermal Amplification Products
The first product of the LAMP reaction was dumbbell-shaped amplicon with a molecular weight of 135 bp (Figure 2).
4.2. Determination of Analytical Specificity ureC Primers for Helicobacter pylori
The specificity of the designed primers was evaluated with pure H. pylori DNA and several other bacterial species, which are commonly present in the gastric biopsy specimens. The tested species were Streptococcus spp., Bacillus spp., Escherichia coli, Citrobacter spp., Klebsiella spp., Enterobacter ATCC 29212, Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis. The LAMP reaction was positive only with H. pylori DNA and there was no observable white turbidity or ladder- like pattern in agarose gel electrophoresis with non-H. pylori bacteria. Also, the PCR reaction was only positive with H. pylori DNA and the 214 bp band was not observed with non-H. pylori bacteria in gel electrophoresis (Figure 3).
M, 100 bp DNA ladder; E, empty; lane 1, negative control; E, empty; lane 2, positive control of H. pylori pure DNA; lane 3, Streptococcus spp.; lane 4, Bacillus spp.; lane 5, Escherichia coli; lane 6, Citrobacter spp.; lane 7, Klebsiella spp.; lane 8, Enterobacte ATCC 29212; lane 9, Staphylococcus aureus ATCC 25923; lane 10, Staphylococcus epidermidis.
4.3. Determination of Analytical Detection Limit for Loop-Mediated Isothermal Amplification
The limit of detection of PCR and LAMP for detection of H. pylori was assessed using tenfold serial dilution of pure H. pylori DNA. The lowest concentration of pure H. pylori DNA that yields positive reaction by LAMP and 214 bp band in PCR was 10fg per reaction. Since the average genome size of the H. pylori DNA is estimated 1.67 × 106 bp, thus, limit of detection in both methods is equivalent to approximately 6 copy numbers of H. pylori DNA in each reaction.
5. Discussion
Helicobacter pylori is very common and approximately 50% of the world’s population is infected by this bacterium. Considering to the fact that the worldwide spread of H. pylori infection and its associated diseases such as gastric cancer and on the other hand since it is possible to decrease risk of gastric cancer with treatment of H. pylori infection, a quick and accurate diagnosis of H. pylori infection is crucial (24). Different methods with different sensitivities are used for diagnosis of H. pylori infections in suspected patients such as histopathology, the rapid urease test, culture, fecal antigen test, serology and molecular methods like PCR and real-time PCR. Since H. pylori is not distributed evenly within the stomach tissue, bacteriological culture of the H. pylori from biopsy specimens has the least amount of sensitivity. In addition, the rapid urease test will also be affected by this condition; so, sensitivity of the test is low (1, 25). As a result, molecular methods are more important when we wish to diagnose H. pylori more accurately. The purpose of the current study was designing, analyzing and comparing PCR and LAMP targeted ureC to detect H. pylori.
The limit of detection obtained in PCR and LAMP were less than the previous reported methods (2, 26, 27). The analytical sensitivity of PCR identified 10 fg of H. pylori DNA per each reaction. Since the H. pylori genome weighs about 1.67 fg the amount of this DNA is equal to 6 copy numbers of H. pylori. This means the sensitivity of PCR, using the primers designed in this research, is 8 times more sensitive than LU’s study and, it is also approximately 2 times more sensitive than the research by (Clayton et al. 1992 (2), Linpisarn et al. 2005 (27)). LU’s and Thoreson reported a limit of detection around 5 H. pylori genomes in their research (0.01 pg and 1 fg, respectively) (26, 28). We believe that the major drawback of Thoreson’s research is that they miscalculated the genome weight of H. pylori as one fg. LU’s used 16srRNA as target of amplification, which is less specific in comparison with ureC gene. The primers used in LAMP were designed on the basis of the conserved regions of the ureC gene. The conserved regions of ureC were identified using alignment of some standard strain ureC gene by ClustalW2 software. Although various genes like ureA, SSA, 16SrRNA and ureC are used as targets in molecular detection of H. pylori, ureC gene was considered in this research since it is located in a completely conserved area in the H. pylori genome and the sensitivity and specificity of this gene in identifying H. pylori are more than those of 16srRNA gene (26).
The detection limit of the LAMP with new designed primers was 6 copy numbers of H. pylori DNA in each reaction. It is less than the detection limit reported by Minami et al. in which the possibility of identifying the least amount of H. pylori using LAMP also targeted the ureC gene was equal to 102 CFU/Tube (29). This means that the LAMP is more sensitive in the present research rather than in the mentioned researches and it has identified a fewer number of bacteria (30-39). Although the obtained results from LAMP absolutely conformed to that of the PCR method in this research and the numbers of bacteria identified were similar in both methods, the LAMP is preferred. The LAMP could be carried out in most of the hospital laboratories even with mediocre facilities as a simple, fast, efficient and easy method. Therefore, it is considered an ideal method for specific identification of H. pylori among all existing bacteria in the stomach and biopsy sample and it is suggested as a highly specific method as well.
The specificity of the method depends on 3 pairs of primers, which identify 8 separate areas of the target gene while in PCR one pair of primers is used. This method can be done in 1.5 hours while in order to perform the PCR method 3 - 4 hours time is needed. The PCR method is also highly specific in identifying H. pylori; however, this method suffers limitations such as needing expensive equipment, having a complex protocol and also need to a thermo cycler machine in order to regulate a cycle of temperatures while the LAMP can be carried out in a isothermal temperature without the need for thermo cycler and expensive equipment. As a result LAMP with the target sequence of the ureC gene is advised as a technique which is highly specialized and sensitive, quick and simple in identifying H. pylori. Therefore, this method could be used as a valuable potential method in clinics and as a routine test in laboratories, although our results are still preliminary and clinical evaluation of the method should also be performed in future.