1. Background
Helicobacter pylori is a Gram-negative, microaerophilic spiral bacteria that colonize in gastric epithelial tissue and mucus of half of the worlds and 80% of the Iranian population (1, 2). The prevalence of H. pylori infection varies all over the world based on geographic area, age, race and sanitary conditions. Helicobacter pylori are mainly acquired in childhood due to lack of proper hygiene (3). Infection with H. pylori is associated with some gastrointestinal disorders, including chronic gastritis, peptic ulceration, gastric mucosa-associated lymphoid tissue lymphoma, intestinal metaplasia and gastric cancer (4). Despite the susceptibility of H. pylori to many antibiotics in vitro, its eradication is difficult in vivo. This is thought to be due to inactivation of antibiotics in acidic environments of the gastric mucosa (5). Commonly, triple therapy including a proton pump inhibitor (PPI) (omeprazole or pantoprazole) together with two antibiotics such as clarithromycin and metronidazole or amoxicillin is used for treating of H. pylori infection. Also, fluoroquinolones, nitrofurans and rifamycins are used as alternatives (6). Clarithromycin has a key role in triple therapy.
Clarithromycin (6-O-methyl erythromycin) is a macrolide antibiotic that inhibits protein synthesis of bacteria by binding to the 50s subunit of bacterial ribosome. Acid stability and good absorption in gastric mucus layer make it a good choice for H. pylori eradication in comparison to other macrolides (7). Unfortunately, appearance and increasing resistance to clarithromycin among H. pylori isolates has been reported all over the world (8). This phenomenon decreases the efficacy of H. pylori eradication rate, for example in a study, the eradication rate for clarithromycin-susceptible isolates was 90%, whereas it was 36% for clarithromycin-resistant isolates (9). Also, resistance to clarithromycin shows geographical variation (8). For instance the rate of clarithromycin resistance that was reported in the north of Iran (Sari) is greater than once reported in south (Shiraz) of this country (10). Therefore, before clarithromycin prescription in Helicobacter pylori treatment, determination of the local susceptibility pattern is necessary in any region.
This is possible by approved antibiotic susceptibility testing (AST) methods against bacteria, such as agar dilution, E-test and disk diffusion. Agar dilution is considered as the gold standard method, yet it is highly laborious and expensive. Therefore, agar dilution could be replaced by E-test or disk diffusion methods (11, 12). E-test is a simple and easy test for determining minimal inhibitory concentration (MIC) of a variety of antibiotics, however it is more expensive. Molecular mechanism of clarithromycin resistance in H. pylori, is related to point mutations in 23S rRNA, which can effect protein synthesis.
The three most popular point mutations in 23S rRNA are A to G transition at position 2142 (A2142G) or 2143 (A2143G) and less frequently, A2142C (13). Several molecular techniques have been developed for the detection of clarithromycin resistance in H. pylori including, restriction fragment length polymorphism (RFLP), 3’-mismatch reverse primer PCR method (3M-PCR), PCR-oligonucleotide ligation assay (OLA), fluorescent in situ hybridization (FISH), PCR-preferential homoduplex formation assay (PHFA) and real-time PCR hybridization assay (5). Among these methods, real-time PCR is known as a simple, rapid and reproducible method for the detection of mutations (14). Many investigators used this method for detection of different locations of point mutations in bacterial or eukaryotic genome (15, 16). For instance, Oleastro et al. (17) used real-time the PCR assay to detect point mutations in 23S rRNA of H. pylori and described this method as a rapid, accurate and reliable method for this goal.
2. Objectives
The aims of this study were the determination of the resistance rate to clarithromycin among local H. pylori isolates by the E-test method at first and further determination of the point mutation profile by analysis of the melting curve by the real-time PCR method.
3. Patients and Methods
3.1. Patients and Bacterial Strains
The sample size (80 biopsies) was determined based on the frequency of resistance to clarithromycin among H. pylori isolates (28%) as reported by the study of Malekzadeh et al. (18). Continuously, from patients with gastrointestinal disorders, who were candidates for endoscopy at Milad and Fayyazbakhsh hospitals (Tehran and Karaj, Iran), three biopsies were taken during 2011 - 2012. One of them was used for rapid urease test and the two remaining were transported to microbial laboratory in Stuart’s transport medium (Merck, Germany) as soon as possible (less than five hours). Biopsies were homogenized, cultured in selective medium, brucella blood agar (Merck, Germany), including vancomycin (Sigma-Aldrich, USA), trimethoprim sulfametoxazol (Sigma-Aldrich, USA), polymyxin B (Sigma-Aldrich, USA) antibiotics, supplemented with sodium pyruvate (Sigma-Aldrich, USA) and ferrous sulfate (Sigma-Aldrich, USA), and then incubated under microaerophilic conditions using the gaspak incubation method (113829 Anaerocult® A, Merck, Type C Gas-Pak) for 48 - 72 hours at 37°C. After appearance of colonies, identification tests including Gram staining, catalase and oxidase tests were done. Also, H. pylori isolates were confirmed by PCR amplification of ureC gene after DNA extraction with the boiling method. Primer sequences (Bioneer company, Korea) are showed in Table 1 (17). This research was accepted by the ethics committee of Shahid Beheshti University of Medical Sciences (SBMU) at 13 Jan 2010.
| Primer/Probe Name | Sequence | Hybridization Region |
|---|---|---|
| ureC (Bioneer, Korea) | 5'-GGATAAGCTTTTAGGGGTGTTAGGGG-3' | |
| 5'-GCTTACTTTCTAACACTAACGCGC-3' | ||
| HPY-S, HPY-A (Bioneer, Korea) | 5'-AGGTTAAGAGGATGCGTCAGTC-3' | |
| 5'-CGCATGATATTCCCATTAGCAGT-3' | ||
| Sensor probe, Anchor probe (Bioneer, Korea) | 5'Cy5-GGCAAGACGGAAAGACC-3' | 2504 - 2520 |
| TGTAGTGGAGGTGAAAATTCCTCCTACCC-3'FAM | 2473 - 2501 |
3.2. Antibiotic Susceptibility Test
Resistance to clarithromycin was tested with epsilometer strips (E-test, Lioflichem Co, Denmark), according to the CLSI protocol and manufacturer’s instructions. Fresh H. pylori isolates were subcultured on selective brucella blood agar (Merck, Germany) for 72 hours. Bacterial suspension equivalent to McFarland turbidity standard three were prepared and spread on non-selective brucella blood agar plates (without antibiotics). After drying of plates, a single E-test strip was placed on them. The plates were incubated under microaerophilic conditions (116275/Anaerocult® C Merck, Germany) at 37°C. After 72 hours, susceptibility and MIC of the isolates were determined, simultaneously. The H. pylori OC1096 strain (bestowed by research center for gastroenterology and hepatology disease, Shahid Beheshti University of Medical Sciences Tehran, Iran) was used as a control isolate. The sensitive (S) and resistant (R) isolates were determined based on the MIC as follow; S ≤ 0.25 mg/L and R ≥ 0.5 mg/L.
3.3. DNA Extraction
DNA extraction was done by the boiling method (19, 20). The concentration of extracted DNA was determined after optical density (OD) detection by the nanodrop instrument (Biointellectic TM Nano 200, Canada).
3.4. UreC Amplification
For confirmation of detected colonies as H. pylori, existence of ureC gene was done by the PCR method. The PCR reaction was carried out in a thermal cycler (Eppendorph Master cycle, Germany) as follow; 35 amplification cycles consisting of a denaturation step at 94°C for one minute, an annealing step at 54°C for one minute and extension step at 72°C for one minute, which were performed after a pre-denaturation of the target DNA at 94°C for four minutes. The final cycle was an extension step at 72°C for five minutes.
3.5. Detection of 23S rRNA Gene Mutations of H. pylori by Real-Time-Polymerase Chain Reaction
The molecular mechanism of clarithromycin resistance was investigated by the real time-PCR method and determination of the melting temperature. Real-time PCR was performed on DNA extracted from H. pylori isolates in Rotor-Gene 6,000 (Corbett). A primer pair (HPY-S and HPY-A, Table 1) was used to amplify the 267 bp fragment of 23s RNA gene. Also, sensor and anchor probes were used to determine point mutations. Real-time PCR assays were performed as follows: an initial denaturation step at 95°C for 10 minutes, followed by 50 cycles with a temperature transition rate of 20°C/second, consisting of 95°C/second, annealing at 60°C for 10 seconds, and extension at 72°C for 17 seconds. After amplification, a melting step was carried out: 95°C/second, cooling to 45°C for 30 seconds with a temperature transition rate of 20°C/second, and finally a slow heating to 85°C at a rate of 0.1°C/second with continuous acquisition of fluorescence decline. Further sequencing of six H. pylori isolates (four resistant, one sensitive and a control strain) was done by the Bioneer Company, Korea.
4. Results
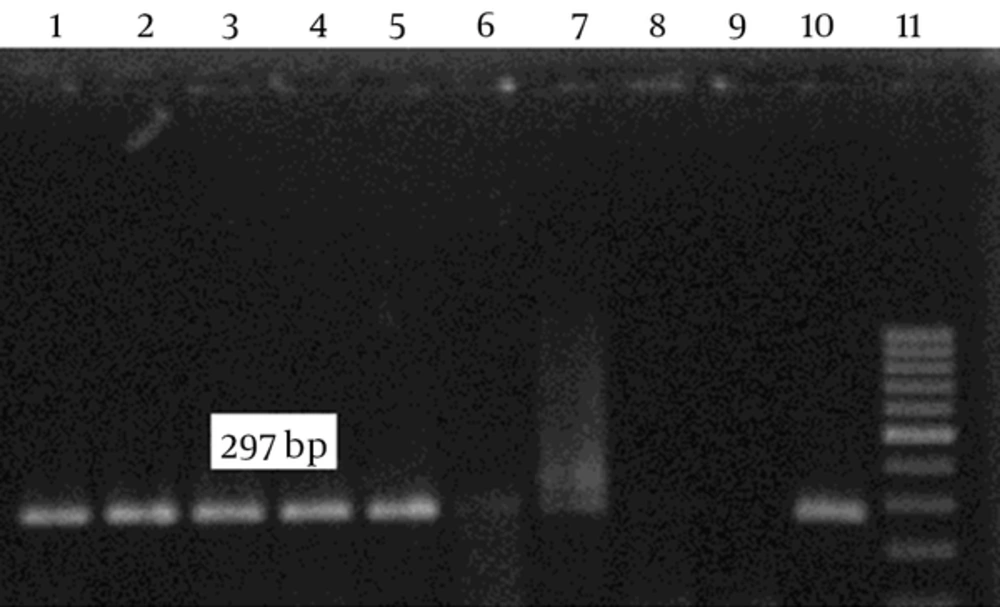
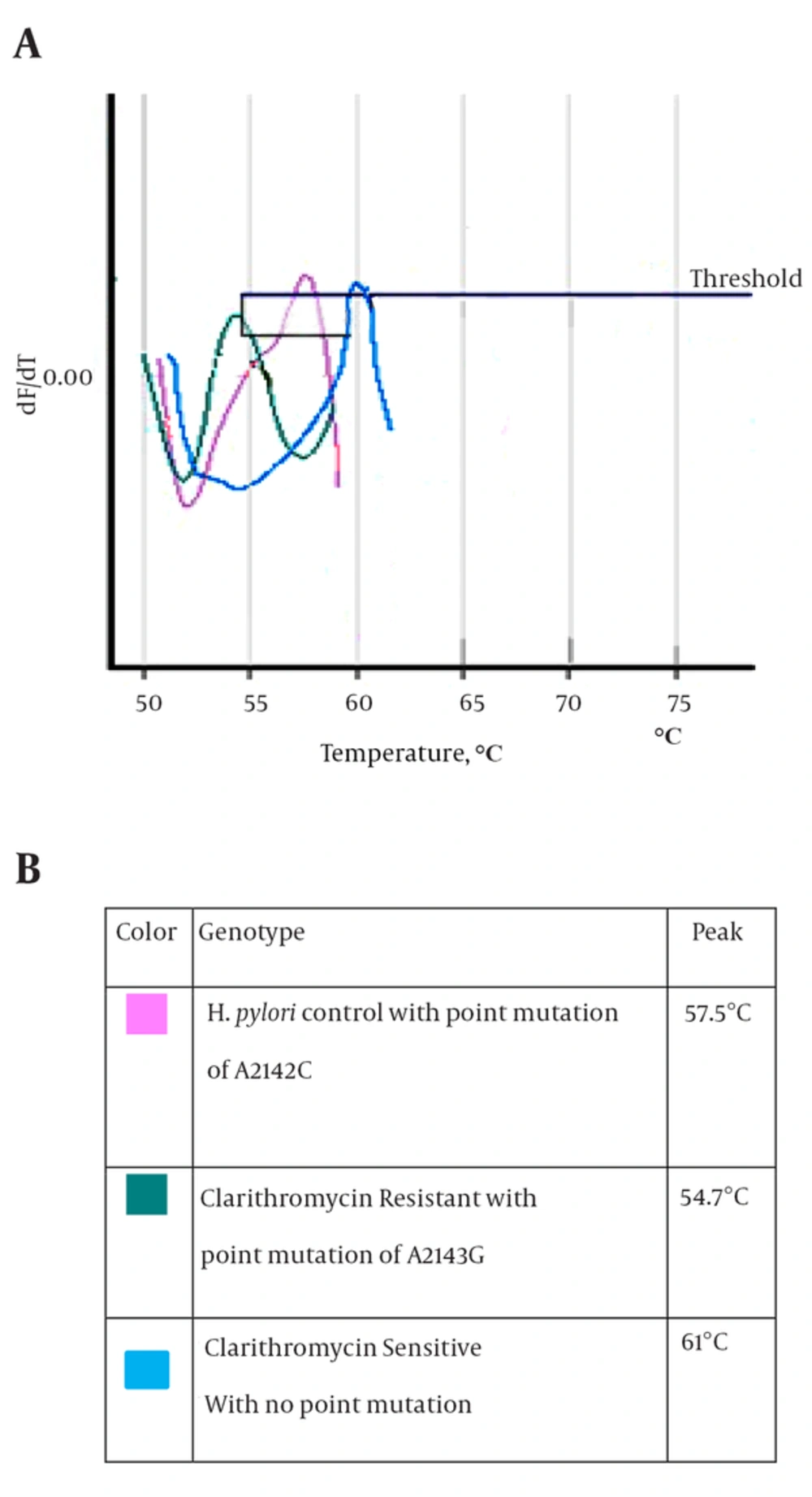
After culture of 80 biopsies, 20 H. pylori strains were isolated (25%). All biochemical tests including Gram staining, catalase and oxidase tests confirmed H. pylori isolates. Also, the presence of ureC gene in all 20 isolates was confirmed by PCR (Figure 1). Resistance to clarithromycin was observed in 4 of 20 H. pylori isolates (21.7%) according to the CLSI standards and by the E-test method. Resistant isolates had MIC of > 0.5 mg/mL (Figure 2). The type of mutation in clarithromycin resistance isolates was determined by real-time PCR. According to Figure 3 melting temperature of probes in susceptible isolates and H. pylori OC1096 strain, was 57.5°C, but in resistance isolates this was 54.7°C. Also, as mentioned before for confirmation of the real-time PCR results, sequencing was done on PCR products of six isolates in parallel. All resistant isolates in this study showed a point mutation at A2143G based on the detected melting temperature and sequencing. Two sequences of amplicons were submitted to NCBI with accession No KP09939 and KP09939.1.
5. Discussion
Helicobacter pylori, is a major cause of upper gastrointestinal disorders such as peptic and duodenal ulcers and gastric cancer. About half of the world’s population is infected with H. pylori (1). One of the best antibiotics that are used in first line therapy for this infection is clarithromycin.
Clarithromycin is a macrolide antibiotic that inhibits protein synthesis of bacteria by binding to the 50 second subunit of bacterial ribosome and preventing of peptidyl transferase activity (21). However, emerging and spreading of antibiotic resistance, especially resistance to clarithromycin, decreases the efficacy of H. pylori infection treatment (22). Therefore, detection of the local antibiotic resistance profile is important. In our study we used the E-test method for determination of resistance to clarithromycin among H. pylori isolates.
Clarithromycin resistance was observed in four of 20 H. pylori isolates (21.7%). Because of high precision and reproducibility of E-test results, we used this method in our study. Similar to our results, the rate of clarithromycin resistance that had been reported by Ogata et al. (23) from Brazil and De Francesco et al. (24) from Italy by the E-test method, was about 22.62% and 18.49%, respectively. However, the rates of clarithromycin resistance that have been described by other studies (by the E-test method) were different. For instance, it was 41.9% in Turkey (25), 35.6% in Spain (26), 5.3% in Costa Rica (27) and 10.6% in Taiwan (28). Existence of such differences is logical, because antibiotic susceptibility profile shows geographical variation between and within countries based on age, race, population density and availability of antibiotics (29). Such discrepancies exist between studies from different parts of Iran. For example, Sirous et al. (30) reported that all H. pylori strains isolated from Iran were sensitive to clarithromycin (by the disk diffusion method). Also, clarithromycin resistance rate among H. pylori isolates in Tehran (by the disk diffusion method) and Southern Iran (Shiraz, by the E-test method) was inconsiderable (4.16% and 5%, respectively) (31, 32). However, other Iranian studies showed considerable rate of clarithromycin resistance among H. pylori isolates e.g. 16% and 17.7% in north west of Iran (Tabriz, by the E-test and modified disk diffusion method) (33, 34), 30% in the north (Sari, by the E-test method) (35), 32% in the west (Ilam, by the disk diffusion method) (36) and 32.4% in Tehran (by the agar dilution method) (37).
The difference between the rates of clarithromycin resistance in studies, which have been done in different parts of Iran may be due to time differences, use of different susceptibility methods, sample sizes, fresh or freeze biopsies and incubation conditions. Over all, this study and other studies that reported high rate of resistance to clarithromycin among H. pylori isolates, showed that clarithromycin resistance rate has reached alarming rates in our country, Iran. Clarithromycin is an expensive antibiotic and not commonly used in Iran, so these rates of resistance could be related to cross-reactivity with other macrolides. For example, Fallahi and Maleknejad (31) reported that the rate of resistance to erythromycin (other kinds of macrolide antibiotics) among H. pylori strains is 4.16%, which is equal to clarithromycin resistance rates.
Resistance to clarithromycin in H. pylori is caused by point mutations at different locations or different base substitutions of 23S rRNA. The most common mutations are A2143G and A2142G, and less frequently A2142C (13). The A2142G and A2142C mutations are associated with high-level cross-resistance to all macrolides; also the A2143G mutation is linked to high-level resistance to erythromycin (38). In this study, we used the real-time PCR method to determine the type of mutation in resistant isolates. First, the 267-bp fragment of 23S rRNA was amplified by Corbett 6,000, and then the mutation was determined by melting curve analysis. According to the melting curve Figure 3, the melting temperature of all resistant H. pylori isolates was at 54.7°C. Therefore, all resistant H. pylori isolates had the A2143G mutation. Also, sequencing results confirmed A2143G mutation. The melting temperature of susceptible and OC1096 strain, were 61ºC and 57.5º C, respectively. Oleastro et al. (17) used the Light thermo cycler apparatus and reported that the melting temperature of susceptible, standard and resistance (A2143G mutation) isolates were 61.5ºC, 58ºC and 53.6ºC, respectively.
The inconsiderable difference to our results (57.6ºC) may be due to the use of different instruments and standard strains. Sadeghifard et al. (36) investigated mutations of 23S rRNA gene among H. pylori isolates by the PCR-restriction fragment length polymorphism (RFLP) method. In that study, all resistant isolates had the A2143G mutation. Also, in a study performed by Kargar et al. (39) using PCR-RFLP, mutation of A2143G was the dominant point mutation among H. pylori isolates. Mohammadi et al. (40) used the PCR-RFLP method and reported 73.68% of H. pylori isolates had the A2143G mutation. All mentioned studies reported similar point mutations among Iranian H. pylori strains despite the use of different methods. Therefore, it seems that the frequency of A2143G mutation among clarithromycin resistant H. pylori isolates from Iranian patients is higher than other mutations. Similar results were observed in clarithromycin-resistant H. pylori strains isolated from Korea (by PCR amplification and nucleotide sequence analyses) (41), Argentina (by PCR-RFLP method) (42) and Colombia (by the PCR-RFLP method) (43). However, other kinds of point mutations of clarithromycin resistance were shown in other studies from Iran. For example, the incidence of A2142G genotype in the study of Naserpour Farivar et al. (44), by scorpion real-time PCR, was higher than other mutations. Also, in the study of Khademi et al. (45) all clarithromycin-resistant H. pylori isolates had the T2243C mutation by PCR and further sequencing methods.
In conclusion, our results and other studies in Iran, despite some controversial results, showed that the rate of clarithromycin resistance is increasing in our country. Also it seems the most popular pattern of clarithromycin resistance among Iranian H. pylori isolates is the A2143G mutation. Also determination of the antibiotic resistance profile and finding alternative treatment regimens in any region of the world is necessary. Finally, real-time PCR is a rapid and reproducible method for detecting mutations in 23S rRNA of H. pylori isolates. For better evaluation of clarithromycin resistance rate and finding the predominant molecular pattern of resistance among Iranian H. pylori isolates, doing such study with a greater sample size is recommended.