1. Background
Helicobacter pylori, a microaerophilic flagellated pathogen with the architecture of Gram-negative bacteria, is a gastric colonizer and a common etiological agent of duodenal and stomach ulcers, accounting for 90% and 80% of these ulcers, respectively. Helicobacter pylori-infected individuals are 2 - 6 times more prone to developing mucosa-associated lymphoid tissue (MALT) lymphoma and gastric cancer (1, 2). Compared to other bacterial pathogens that display high clonality, such as Mycobacterium tuberculosis and Shigella dysenteriae, H. pylori lacks clonality, as it is genetically diverse. Thus, every positive case of H. pylori carries a different strain (3). Genetic heterogeneity plays a key role in H. pylori’s adaptation to the host gastric conditions, as well as in escaping the host’s mediated immune responses. Nucleotide-level diversity occurs via several molecular mechanisms, including variation at the transcription and translation levels and mutation. Slipped strand mispairing is considered the most common mechanism mediating phase variation and occurs in the G or C homopolymeric tract, resulting in changes in the reading frames of the gene, which in turn affects the translation of the affected gene. This leads to reversible phenotypic variation with only minor genotypic variation (4).
The phase-variation process results in reversible ON and OFF switching of selected genes in the bacterial genome, providing the advantage of adaptation to organisms in different sets of conditions. Phase-variation-mediated adaptation aids in the survival of subpopulations more suitable for colonizing in a particular environment compared to neighbor subpopulations in the same environment; as a result, the fittest subpopulation will rise and show dominance via the clonal replacement process. This process has been mostly associated with genes encoding cell-surface-structure proteins, which are mainly responsible for interactions with the host; however, genes of the restriction-modification (R-M) system also exhibit phase-variation phenomena (5).
The complete genome sequencing of H. pylori by the Institute for genomic research (TIGR) was a major contributor to accelerating research on this important bacterial pathogen (6). The slipped strand mispairing mechanism of short tandem repeats in the H. pylori genome plays an important role in its adaptation (5). Tomb et al. (1997) previously reported 17 intragenic repeats in addition to 17 genes associated with intergenic repeats (6). Saunder et al. (1998) found 27 putative phase-variable genes in H. pylori 26695 strains (7). A list of 26 genes with potential phase-variation, seven of which were new, were identified by Alm et al. (1999) via a comparative analysis using the complete genome sequence of another H. pylori strain, J99 (8). Similarly, Tannaes et al. (2001) found a new phase-variable gene for phospholipase A, called pldA (9). Salaun et al. (2003) revealed a list of 46 genes in H. pylori using a comparative analysis of the 26695 and J99 genomes. These genes were systemically investigated via polymerase chain reaction (PCR) for their presence or absence, and were then sequenced to determine their repeat and length variability. The study was carried out with 23 different geographic-based H. pylori strains, including East Asian, Ladakh, European, and African strains, and revealed different distribution patterns for phase-variable genes in these strains, confirming that the patterns vary from region to region (10). Salaun et al. (2004) also conducted an experimentally designed study for phase-variable genes in three unrelated H. pylori strains, including the Sydney strain (SS1), Hp141, and Hp145, and found different distribution patterns of phase-variable genes in these strains (11).
Keeping in view the above points, we conducted the present study to investigate the distribution patterns of phase-variable genes in Pakistani H. pylori strains, and to analyze and compare them with strains prevalent in other regions of the world.
2. Objectives
The present study was conducted with a focus on the following objectives: to determine the prevalence of H. pylori infections based on sex and age, to investigate the distribution patterns of phase-variable genes in Pakistani H. pylori strains, and to analyze and compare them with strains prevalent in other regions of the world.
3. Patients and Methods
3.1. Patients
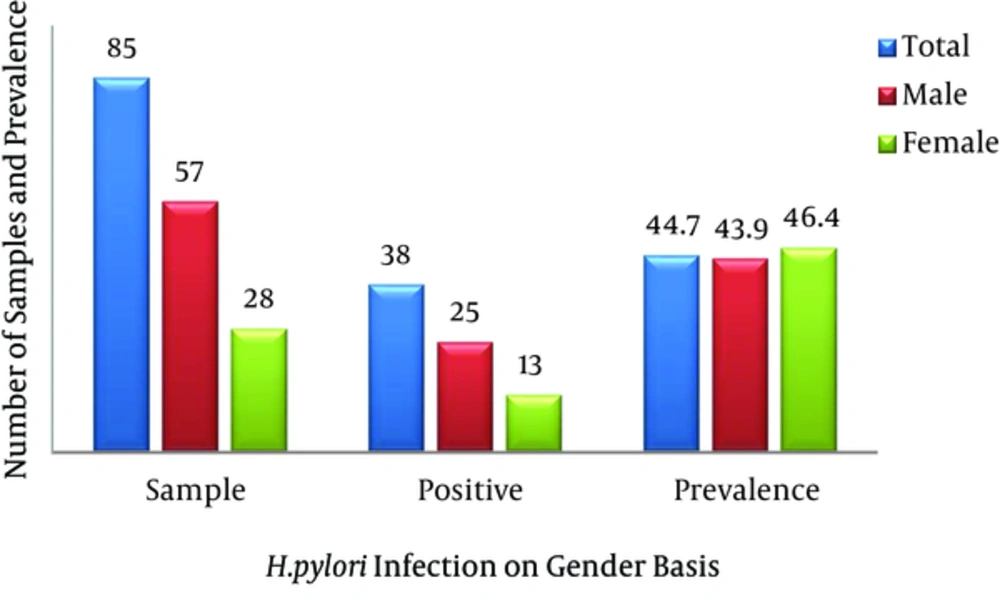
This research was done at the department of microbiology, Quaid-i-Azam University, Islamabad. Biopsy samples from 85 patients were investigated. The patients included in the study were suffering from various upper gastrointestinal tract symptoms, such as nausea, diarrhea, abdominal pain, and heartburn. The patients’ ages were in the range of 10 - 85 years, and the population consisted of 57 males and 28 females. All patients underwent endoscopy at the gastrointestinal endoscopy department, military hospital, Rawalpindi, after giving informed consent. The protocol was approved by the hospital’s ethics committee.
3.2. Culturing
Biopsy specimens were collected from each patient and dipped into a 20% glucose solution for culturing purposes (12). Gastric biopsy specimens were then chopped and inoculated on Columbia blood agar media (CM0331B; Oxoid) supplemented with 7% laked horse blood (SR0048; Oxoid), which also contained Dent’s H. pylori selective medium (SR0147E; Oxoid). The culturing conditions for H. pylori growth were provided by using a Campylobacter gas-generating kit (BR0056A, Oxoid) in which a microaerophilic environment, i.e. 10% carbon dioxide (CO2), 5% oxygen (O2), and 85% nitrogen (N2), was already established. Starting after three days of incubation, the plates were checked on a daily basis for H. pylori growth; if no growth was observed after seven days, the plate was considered negative for H. pylori. Positive cultures were in the form of small rounded colonies, which were sub-cultured to obtain pure cultures. Positive isolates of H. pylori were further identified.
3.3. Identification of Helicobacter pylori-Positive Samples
Identification of positive isolates was done using Gram staining and catalase, oxidase, and urease tests (13).
3.4. Genomic DNA Isolation
Genomic DNA extraction from all positive H. pylori isolates was done using ethanol precipitation and the proteinase K treatment method with some modifications not described in previous studies (14). A single colony of bacteria was chosen and suspended in 20 µl of sodium dodecyl sulfate (1%), 40 µL of proteinase K (100 g/mL), and 80 µL of proteinase buffer (4 M NaCl and 0.5 M EDTA; pH 7.5). Following incubation for 1 hour at 55°C, 100 µL of NaCl (6 M) was added. The sample was vortexed for approximately 1 minute, then centrifuged (16,8639 ×g at 4°C for 1 minute). The addition of absolute ethanol resulted in nucleic acid precipitation, which was then centrifuged for harvest using the abovementioned conditions. Pellets containing DNA formed and were washed with 70% (v/v) ethanol. The resultant product was then suspended in 100 µL of Tris (10 mM) and EDTA buffer (1 mM), keeping the pH at 7.5. The final product was then stored at -20°C before PCR analysis.
3.5. Primer Designing
Primer was designed using OligoCalc, an oligonucleotide properties calculator (version 3.26), for five phase-variable genes harboring homopolymeric tracts of G and C nucleotides. The primers against each gene were designed around the flanking regions of the repeat tracts located within the open reading frame of the gene (see Appendix 1 in supplementary file). For this purpose, the FASTA format of the gene sequence was downloaded from the national center for biotechnology information (NCBI) and the primers were designed according to the stipulated standard parameters given in the literature.
3.6. Control Strain
One control strain of H. pylori G27 was used in the current study. The control strain was provided by the department of microbiology, QAU, and was tested via PCR to check primer specificity.
3.7. PCR Conditions for Gene Amplification
All PCR reagents were supplied by Fermentas thermo scientific (USA) and primers were obtained from e-Oligo (Hawthorne, NY, USA). Each PCR reaction contained 10X PCR buffer, MgCl2 (25 mM), dNTPs (10 mM), primer (25 pm), Taq polymerase (5 U) and 5 µL of template DNA.
3.8. Composition of PCR Reaction Mixture
The reaction mixture was prepared by using 5 µL of DNA sample, 2.5 µL of 10 X buffer, 1.5 µL of MgCl2, 0.4 µL of dNTPs, 0.3 µL of Taq polymerase, and 0.4 µL each of forward and reverse primer; the remaining volume was filled by 14.5 µL of PCR water to make a final volume of 25 µL (see Appendix 2 in supplementary file).
3.9. Agarose Gel Electrophoresis for Identification of PCR Products
PCR for five phase-variable genes yielded products that were separated, and specific PCR products were analyzed via the 2% gel electrophoresis process. The gel was run at 110 V for 40 - 45 minutes depending on the desired separation of the PCR product in the gel electrophoresis apparatus (Whatman BioProducts). The gel was then placed under UV light in the gel Doc system for visualization. The results were noted and the interpretation of the gel was done according to the size of the desired product.
3.10. Statistical Analysis
Statistical analysis was carried out using SPSS software, version 16 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
4. Results
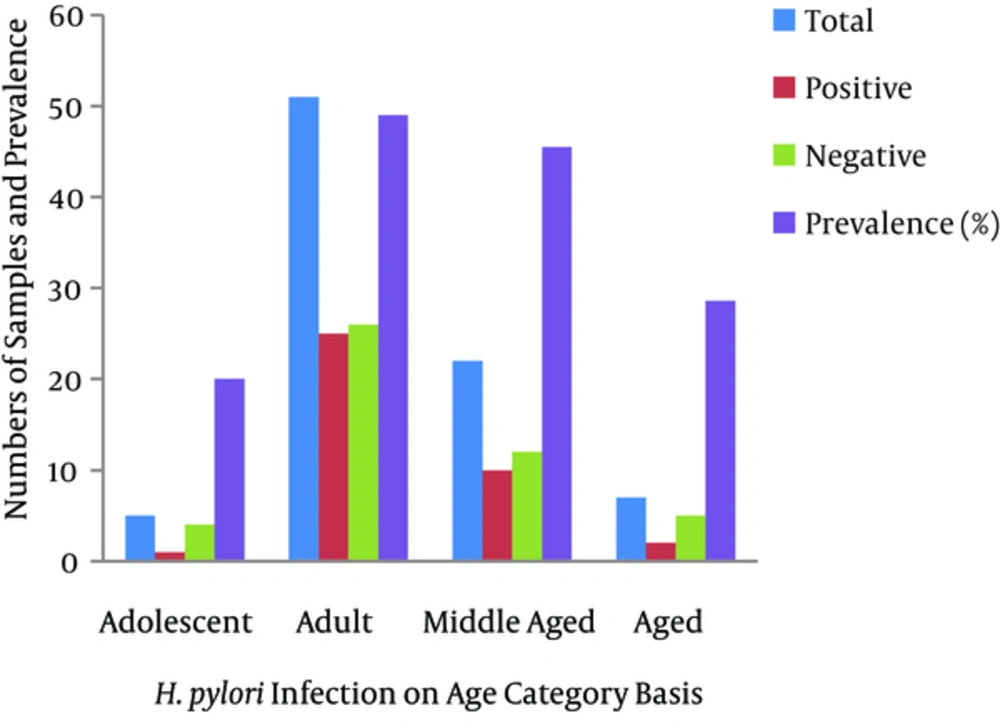
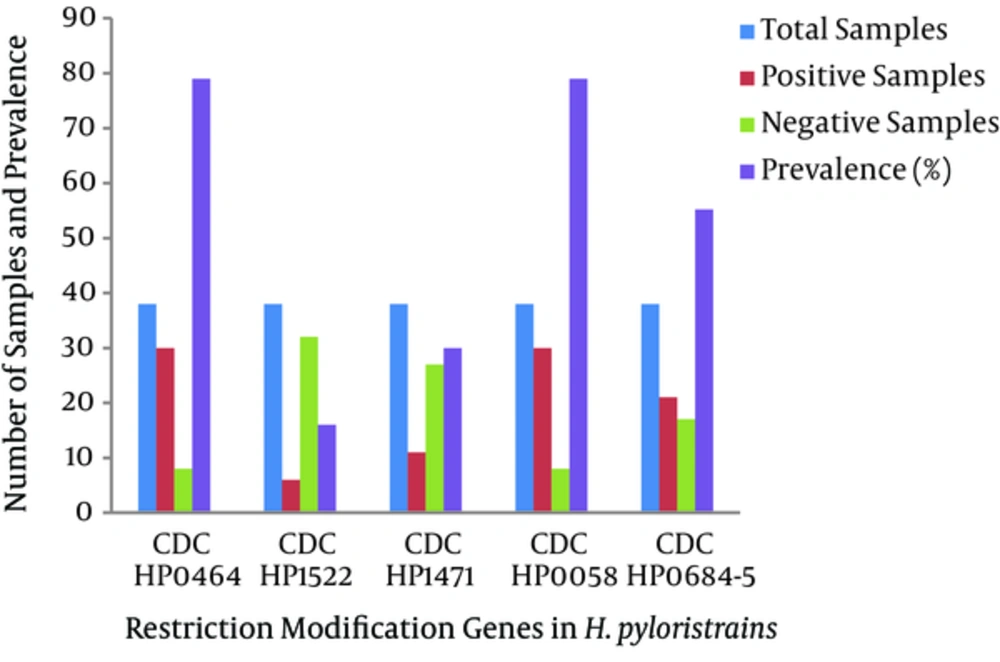
Eighty-five samples were used in the current study, of which 57 were from males and 28 were from females. There were five adolescent patients (age range 13 - 28 years), 51 adults (19 - 44 years), 22 middle-aged patients (45 - 64 years), and seven elderly patients (> 60 years). Twenty-five (43.9%) of the 42 male biopsy samples had positive cultures, while 13 of the 28 (46.4%) female samples were positive. Cultures were positive for one (20%) of the adolescents, 25 (69%) of the young adults, 10 (45.5%) of the middle-aged patients, and two (28.6%) of the elderly. The dominance of the type I R-M system was shown by the 79% prevalence of hsdR, the gene encoding the restriction enzyme of the type I R-M system. This was followed by the type II R-M system, with 30% prevalence of the β-subunit of the restriction enzyme. The type III system was the least prevalent in the Pakistani H. pylori strains, as mod, the gene encoding the methyltransferase enzyme, was revealed in 16% of the samples. Our study also revealed a high prevalence of the streptococcal M protein (79%) gene and the fliP gene (56%) in Pakistani H. pylori strains.
| Feature | Patients (n) | H. pylori-positive (%) | Odds ratio (95% CL) | P Value |
|---|---|---|---|---|
| Status | 85 | 38 (44.7) | ||
| Gender | 0.823 | |||
| Male | 57 | 25 (43.9) | 1.00 (1.00 - 1.00) | |
| Female | 28 | 13 (46.4 ) | 0.90 (0.36 - 2.23) | |
| Age category | ||||
| Adolescent (13 - 18) | 5 | 1 (20.0) | 1.00 (1.00 - 1.00) | 1.000 |
| Adult (19 - 44) | 51 | 25 (49.0) | 0.26 (0.02 - 2.48) | 0.214 |
| Middle-aged (45 - 64) | 22 | 10 (45.5) | 0.30 (0.02 - 3.13) | 0.296 |
| Elderly (> 65) | 7 | 2 (28.6) | 0.62 (0.04 - 9.65) | 0.735 |
4.1. Amplification of the hsdR Gene, a type I R-M System Restriction Enzyme (Gene CDC: HP0464)
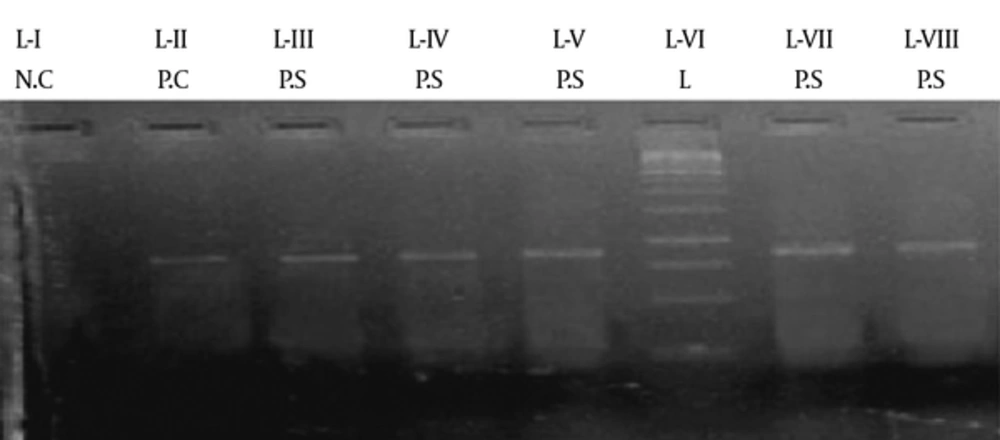
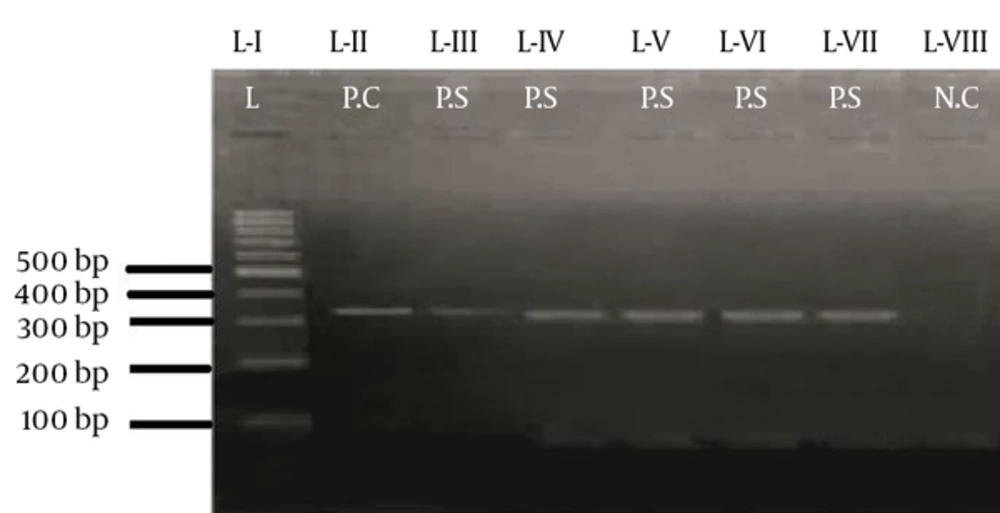
PCR was performed using the program mentioned in Appendix 3 (See Supplementary File). Thirty of 38 samples (79%) were positive, while nine (21%) did not reveal the presence of this gene. Using the primer mentioned in Appendix 1 (supplementary file) the region of 872 bp was amplified and recognized with the help of a ladder of 1 kb. A positive control was used in the form of the G27 strain, while the negative control did not contain template DNA. The gel-documented picture of the amplified gene along with positive and negative samples plus the 1 kb ladder is shown in Figure 3.
4.2. Amplification of β-Subunit of the Restriction Enzyme of the Type II R-M System (Gene CDC: HP1471)
The gene was amplified using the primers mentioned in Appendix 1 (see supplementary file). Eleven samples showed the presence of the gene, while 27 did not. The size of the desired gene was 400 bp, which was recognized using a 100 bp ladder. The H. pylori G27 strain was used as the positive control, while the negative control lacked template DNA. The PCR profile for the mentioned gene is given in Appendix 4 (see supplementary file). The gel representation for the gene is shown in Figure 4.
4.3. Amplification of the Mod Gene of the Type III R-M System (Gene CDC: HP1522)
The region of 330 bp was amplified using the PCR profile mentioned in Appendix 5 (see supplementary file). Seven of 39 samples (16%) revealed the presence of the gene and 84% did not. The size of the desired gene was determined using a ladder of 100 bp. The positive control was in the form of the G27 strain, while the negative control lacked template DNA. The primers used for this gene amplification are mentioned in Appendix 1 (see supplementary file).The gel-documented picture of this gene, along with the positive and negative controls plus the 1 kb ladder, is shown in Figure 5.
4.4. Molecular Detection of fliP Gene (Gene CDC: HP0684-5)
Using primers, a region of 400 bp of the fliP gene was amplified. Out of 38 DNA samples, 21 (55.2%) were positive for this gene, while it was not detected in 17 (44.8%) samples.
4.5. Molecular Detection of Streptococcal M Protein Gene (Gene CDC: HP0085)
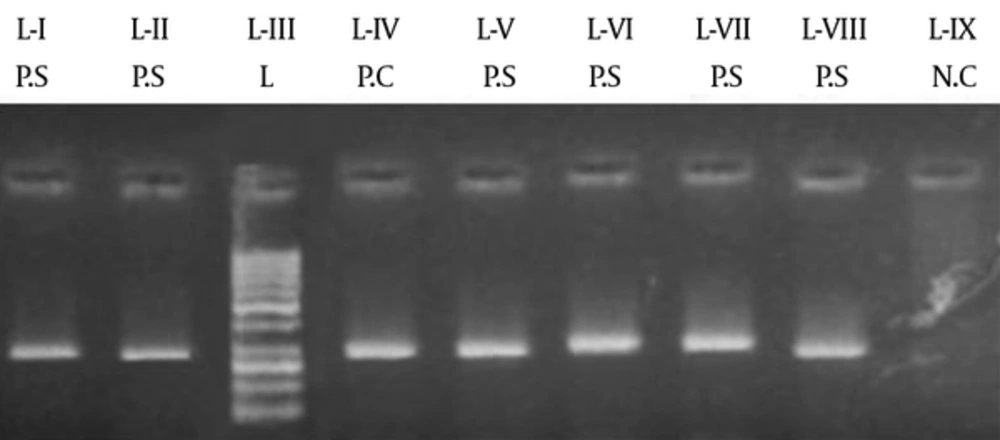
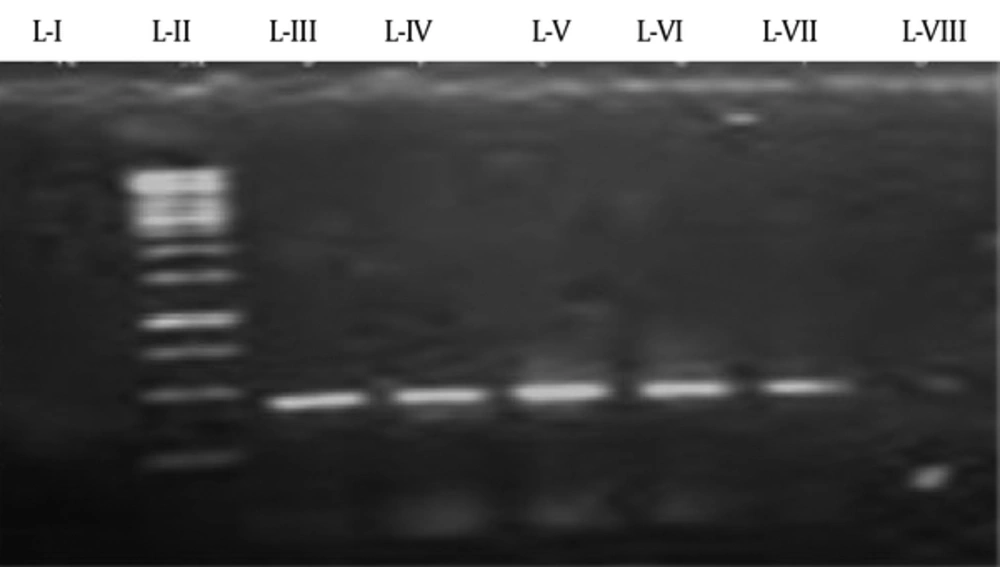
A region of 900 bp was amplified for streptococcal M protein gene. Number of positive and negative samples can be seen in Figure 8.Figure 7.
L-I shows the negative control. L-II, L-III, L-IV, L-V, and L-VI show positive samples, while L-VII shows the marker of 1 kb and L-VIII shows the positive control of G27.
4.6. Prevalence of Phase-Variable Genes in Pakistani Helicobacter Pylori Strains
The HP0464 gene was present in 79% of the samples, while only 21% showed negative results, indicating the presence of this gene in the majority of strains infecting the population. However, HP1522 and HP1471 were present in 16% and 30%, respectively, of the bacterial populations infecting the patients, revealing the lesser role of these genes compared to HP0464 in the prevalence and propagation of H. pylori strains in Pakistan. Similarly, the streptococcal M protein gene also exhibited a high prevalence in Pakistani clinical H. pylori strains, indicating their role in increased virulence.
5. Discussion
The current study was conducted to investigate the prevalence of H. pylori infections in the study population according to sex and age. The study was also designed to investigate the presence or absence of phase-variable genes in H. pylori strains present in Pakistan, which have already been reported in European strains of H. pylori. The epidemiology of the phase-variability differs from strain to strain and from region to region. This investigation helped to clarify the distribution patterns of phase-variable genes of R-M systems in H. pylori strains of Pakistan, and compared them to H. pylori strains in the other parts of world, especially Europe.
Rasheed et al. (2011) revealed a 32.7% prevalence of H. pylori infections in 110 symptomatic patients. That study included 66.4% males and 33.6% females in the age range of 10 - 85 years. On cultures, it was found that 37% of the males and 24.3% of the females were infected. The incidence of infection was high in young adults (34.3%), middle-aged patients (32.1%), and the elderly (28.6%) compared to adolescents. No statistical difference was observed between the sexes or based on age (14). We found a high prevalence of H. pylori infections in the studied population by culturing (44.7%, higher than previous findings). Our findings revealed a higher prevalence in females (46.4%) than in males (43.9%), opposite to the abovementioned findings. Similar to previous findings, we found similar distribution patterns of H. pylori infections in young adults (49.0%), middle-aged patients (45.5%), and the elderly (28.6%), confirming that H. pylori infections increase with age. The probability value was found to be statistically non-significant based on both gender and age.
A prevalence study of H. pylori infections was carried out in the population of Barakahu, Islamabad, from 2008 to 2009 and reported a 74.4% H. pylori infection rate. It was revealed that H. pylori infections were more prevalent in females (75.4%) than in males (73.5%), and that they were highly prevalent in adults (81%), increasing with age (15). In the present study, H. pylori infections were reported in 44.4% of the study population. A high prevalence of H. pylori infection was found in young adults (29%) and the middle-aged (45.5%). The overall reported prevalence was very high compared to ours, which may be due to sample sizes, the methods by which cultures were considered positive, and the correlation of different risk factors.
Previous research reported a list of 46 putative phase-variable genes in the H. pylori genome, including nine of the R-M group. Our investigation confirmed the prevalence of five phase-variable genes in Pakistani H. pylori strains, and revealed the dominance of the type I R-M system with a 79% prevalence of hsdR, the gene encoding the restriction enzyme of the type I R-M system. This was followed by the type II R-M system, with a 30% prevalence of the β-subunit of the restriction enzyme. The type III system was the least prevalent in Pakistani H. pylori strains, as mod, the gene encoding the methyltransferase enzyme, was revealed in 16% of the samples. Our results also confirmed the presence of the fliP (56%) and SMP (79%) genes in H. pylori strains isolated from patients presenting at military hospital, Rawalpindi.
The story of phase-variable genes in H. pylori began in 1997 when Tomb et al. published the complete genome sequence of H. pylori strain 26695 (6). All three types of R-M system genes were reported, results that were confirmed by later studies. Similarly, a sequence analysis of the J99 Hp strain revealed the presence of all three R-M system genes (8). A similar distribution pattern of the hsdR gene was found in all European strains, as well as in African and Ladakh H. pylori strains, showing the dominance of the type I R-M system. The type III R-M system mod gene was found in only three European strains, 26695, SS1, and VZ21, and was absent in B225, H1413, NQ367, 111UK, and 105UK. In addition, this gene is present in the JP96-9 Hp strain, but absent in the remaining four East Asian H. pylori strains, GU17, GU5, RE12001, and L133. Moreover, the L7, L72, and L67 strains prevalent in Ladakh lack the mod gene. African strains such as J99 and 162.0 harbor the mod gene, while the remaining strains do not: ISU2003-1, C164, CC31C, CC42C, and 129.0.
The type II system β-subunit of the restriction enzyme was present in all East Asian and Ladakh strains. This gene is present in 26695, B225, and VZ21, but absent in the remaining six European strains. The African strains J99, CC31C, CC42C, and 162.0 harbor this gene, while the remaining three lack it (10). That study revealed the dominance of the hsdR gene in all of the study strains obtained from the different geographical locations, similar to the present study; however, our study suggested that this gene is only present in 79% of Pakistani Hp strains, not in all of the study strains, which gives a clue about the possibilities of newly evolved strains present in the Pakistani population. The dominance of one R-M system in Pakistani Hp strains helps these strains to better invade both micro- and macroenvironments, and make their host more resistant and fittest subpopulation.
A somewhat similar distribution pattern of the mod gene was found in the present investigation compared to its prevalence in Asian strains; however, in European strains it was a bit more prevalent than in our study. The Ladakh strains were found to completely lack this gene, while the African strains revealed a lower prevalence, which was similar to our findings. The type II R-M system β-subunit of the restriction enzyme was found in only 30% of Pakistani Hp strains, findings that are opposite to previous findings in Asian and Ladakh strains, which identified this gene in all studied Hp strains. This finding also suggests the possibility of newly evolved strains with different configurations of R-M system genes. A higher prevalence of this gene was reported in African strains than in our findings, while European strains revealed similar distribution patterns.
Salaun et al. (2004) conducted a functional study of putative phase-variable genes of H. pylori in three unrelated European H. pylori strains, SS1, Hp141, and Hp145. The type I R-M system gene, hsdR, was found to be present in all three strains, suggesting their dominance. The type III R-M system mod gene was found to be prevalent in two of the strains, SS1 and Hp145, but absent in the Hp141 strain. Similarly, the type II R-M system gene, the β-subunit of the restriction enzyme, was revealed in Hp141 and Hp145 but absent in SS1 (11). That study also revealed the dominance of the type I R-M system in three European Hp strains, similar to our findings as well as to previous reports. Molecular analysis revealed a 92% prevalence of the type I R-M system hsdR gene in three Hp strains: 26695, J99, and HPAG (16). Molecular investigations of phase-variable R-M genes have also been carried out on the Hp strain OND79, confirming their presence in six out of nine of the mentioned Hp strains, and further functional analyses were conducted. All three types of R-M system genes were reported to be equally distributed in the studied strain, while the functional analysis showed that the mod gene is a main player in colonization and in the regulation of different gene expressions (17).
Salaun et al. (2003) reported a 100% presence of the fliP gene in East Asian, Ladakh, and European strains, with a 50% prevalence in African strains (10). The prevalence of the SMP gene was 62% in European strains, 60% in Asian strains, and 75% in African strains (11). The prevalence of the fliP and SMP genes in the present study was relatively similar to that of African strains. This similar trend could be due to the socioeconomic or hygienic conditions of the populations in either region, or could be attributed to mutations, which possibly play an important role in creating genetic variations in H. pylori. The findings of this study also suggest the presence of simple sequence repeats (SSRs) in the abovementioned phase-variable genes of H. pylori. The fliP and SMP genes basically code for cell-surface-associated proteins, which add to the pathogenicity and motility of H. pylori in the viscous environment of the human stomach. Colonization is the most essential step in the pathogenicity of an organism, and the fliP gene helps in flagellar expression and colonization of H. pylori. The fliP gene codes for a component of flagellar basal body proteins, while the SMP gene helps in immune evasion by resisting phagocytosis.
The current study suggests a high rate of H. pylori infections in females compared to males. Similarly, the incidence of H. pylori infections was found to increase with age. Adult and elderly patients were found to have the most infections with H. pylori. To our knowledge, this is the first study to investigate phase-variable genes in Pakistani H. pylori strains. The dominance of one type of R-M system over others in these strains was an interesting finding, considering their role in bacterial defense in the macroenvironment. Type I R-M systems might allow Pakistani H. pylori strains to better colonize different environmental conditions and different sites of infection, as well to smartly evade host immune responses. The study also revealed the prevalence of phase-variable genes of type II and type III R-M system genes; however, their incidence was lower, showing their smaller role in Hp strain survival. The distribution pattern of the type III R-M system mod gene in comparison to Asian strains was different, an interesting finding that opens up new areas of research on R-M systems in H. pylori strains prevalent in Pakistan. Moreover, the investigated phase-variable R-M genes might be instrumental in generating genetic diversity by changing their phases from ON to OFF or vice versa, thereby increasing colonization and providing overall survival and fitness advantages to H. pylori strains in the Pakistani population.
The present study included a small sample size in terms of positive cultures for H. pylori, and confirmation of specific strains was not done. Further studies that include larger sample sizes from different regions of the country, confirming the specific strains and analyzing the strains based on gender and age will help increase the understanding of the overall picture of resistant H. pylori strains and their pathogenesis. Phase-variable gene profiling has not yet been done in Pakistani strains of H. pylori. This is the first step toward profiling already-reported phase-variable genes in the H. pylori strains circulating in Pakistan. Investigation of the distributions and profiles of these genes is important for understanding the complete pathogenesis of this unusual bacterium, in light of its high prevalence in Pakistani populations. The present study was an important investigation that will help to stimulate research on this important aspect of Pakistani H. pylori strains.
5.1. Conclusion
The varied patterns of distribution of phase-variable genes within the H. pylori strains prevalent in Pakistan, especially the dominant prevalence of type I R-M gene HP0464, was an interesting finding, considering the role of such systems in bacterial defense against phages in the macroenvironment. The dominance of one form of R-M system over another might be able to confer a competitive advantage on H. pylori strains to better invade both macro- and microenvironments. Moreover, the investigated phase-variable genes might be instrumental in generating genetic diversity by changing their phases from ON to OFF or vice versa, thereby increasing colonization and providing overall survival and fitness advantages to H. pylori strains in the Pakistani population.