1. Background
Many species of pathogenic bacteria, including Staphylococcus aureus, have become antibiotic resistant. The potential of S. aureus to develop resistance rapidly to many antibiotics has led to the emergence of methicillin-resistant S. aureus (MRSA) (1, 2). Methicillin resistance is associated with the production of the PBP2a protein by the mecA gene. This gene is located on a 30 - 50 kb chromosomal DNA fragment, which is present in resistant strains but absent in susceptible ones (2, 3). The staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element that carries the mecA gene and other antibiotic-resistant genes in MRSA strains (4, 5). Staphylococcal cassette chromosome mec typing is performed to identify and differentiate community-associated MRSA infections from healthcare-associated ones. Staphylococcal cassette chromosome mec typing has been widely used to identify several subtypes or variants of the main SCCmec types. Staphylococcal cassette chromosome mec typing using the multiplex polymerase chain reaction (PCR) is a simple method that can be applied in clinical microbiology laboratories (6, 7).
Approximately 44% of nosocomial infections worldwide are estimated to be due to MRSA (8). Moreover, MRSA is associated with excessively high healthcare costs in many countries. For example, annual costs to deal with the consequences of MRSA in the U.S. are estimated at over 13.8-billion dollars (9, 10). The prevalence of MRSA continues to increase among healthcare staff, with a high rate of nasal colonization by MRSA reported (11). Methicillin resistant S. aureus can be transmitted via the skin and hands of healthcare staff through the nostrils and lead to infection and a variety of complications in patients (12). MRSA infection can result in severe complications, such as endocarditis, septicemia, pneumonia, and osteomyelitis (13-15).
Restrictions placed on drug treatment due to MRSA have made it more difficult to combat nosocomial and community-acquired infections (16-18). The transmission of MRSA to patients via the hands or nostrils of healthcare staff can result in hospitalized patients experiencing major problems (19). Methicillin resistant S. aureus infections among healthcare staff remain high, and the treatment and hospitalization costs of MRSA-infected patients are enormous. Therefore, strategies to control the spread of MRSA are needed (20). Given the importance of the prevention and treatment of nosocomial infections, data are needed on the incidence rate, prevalence, antibiotic resistance, and bacterial typing of MRSA.
2. Objectives
This study was conducted to determine the frequency, molecular types, and drug resistance of S. aureus nasal isolates in two teaching hospitals in Iran using PCR and disk-diffusion methods.
3. Methods
3.1. Specimen Collection and Bacterial Identification
After obtaining ethical approval for the study from the ethics committee of Shahrekord University of Medical Sciences (Grant No. 1184), 262 people were enrolled in the study: 149 staff from Kashani hospital and 113 staff from Hajar hospital. The occupational categories consisted of physicians, nurses, health workers, technicians, administrative staff, and service personnel from the surgical ward, intensive care unit, kitchen, and laundry room.
To ensure qualitative and quantitative standardization in the study, one individual collected all the samples and recorded the data. The nasal specimens were collected from the anterior nares of the participants using labeled sterile cotton wool swabs. The specimens were immediately transferred to trypticase soy broth medium and incubated at 37°C for 24 hours. The specimens were cultured in blood agar and mannitol salt agar media (Hi Media, India) in the laboratory. If the colony became yellow in the mannitol salt agar medium, gram staining was conducted. To differentiate Staphylococcus spp. in gram-positive cluster-forming cocci, catalase and coagulase (with rabbit plasma) production tests were conducted, as well as DNase activity tests on DNase agar (Hi Media, India) and novobiocin susceptibility tests (21, 22). To detect MRSA isolates, oxacillin (1 µg) disk (Hi Media, India) diffusion testing was performed. To determine the antibiotic susceptibility of the isolates, eight antibiotic disks (gentamicin, linezolid, vancomycin, rifampin, novobiocin, ticoplanin, tigecycline, and quinopristin-dalfopristin) obtained from Hi Media (India) were tested using the Kirby–Bauer disk-diffusion method (23).
3.2. DNA Extraction
Total DNA was extracted from the bacteria that grew on the culture media with a genomic DNA purification kit (CinnaGen Co., Iran, according to the manufacturer’s instructions. The quality of the extracted DNA was measured at 260 nm wavelength according to Sambrook and Russell’s method (24). The extracted DNA was stored at -20° C for later use.
3.3. Multiplex PCR Assay for Assignment of the mec Element Type
A multiplex PCR assay was used to identify the MRSA isolates. The multiplex PCR method is a rapid, accurate, and useful assay to detect the mecA gene in MRSA strains, particularly in a hospital setting (25).
The multiplex PCR included eight loci, A-H, selected based on mec element sequences (Table 1) (26). The mecA gene was also included in this protocol. The PCR reactions were conducted in a total volume of 25 μL containing the following: 2 μL of DNA sample, 2.5 µl of 10× PCR buffer, 2 µL of mixed dNTP, 2.5 µL of MgCl2, 0.5 µL of DNA Taq polymerase, and 0.5 µL of primers A and D for type I; primers B, C, D, and G for type II; primers C, E, F, and H for type III, and primers D and I for type IV. The PCR assay was performed in a DNA Thermal Cycler 480 (Applied Biosystems, U.S.) using the following parameters: denaturation for 5 min at 95°C; 35 cycles of 94°C for 45 seconds, 53°C for 40 seconds, and 72°C for 1 minutes, followed by a final extension for 7 minutes at 72°C. For type IV, the annealing temperature was 52°C for 50 seconds. The PCR products then underwent polyacrylamide gel (8%) electrophoresis and staining with silver nitrate.
| Locus | Primer | Oligonucleotide Sequence (5’ - 3’) | Location | Amplicon Size, bp | Specificity, SCCmec Type |
|---|---|---|---|---|---|
| A | CIF2 F2 | TTCGAGTTGCTGATGAAGAAGG | 18398 - 18419b | 495 | I |
| CIF2 R2 | ATTTACCACAAGGACTACCAGC | 18892 - 18871b | |||
| B | KDP F1 | AATCATCTGCCATTGGTGATGC | 10445 - 10467c | 284 | II |
| KDP R1 | CGAATGAAGTGAAAGAAAGTGG | 10728 - 10707c | |||
| C | MECI P2 | ATCAAGACTTGCATTCAGGC | 42428 - 42447c | 209 | II, III |
| MECI P3 | GCGGTTTCAATTCACTTGTC | 42636 - 42617c | |||
| D | DCS F2 | CATCCTATGATAGCTTGGTC | 38011 - 37992b | 342 | I, II, IV |
| DCS R1 | CTAAATCATAGCCATGACCG | 37670 - 37689b | |||
| E | RIF4 F3 | GTGATTGTTCGAGATATGTGG | 45587 - 45607d | 243 | III |
| RIF4 R9 | CGCTTTATCTGTATCTATCGC | 45829 - 45809d | |||
| F | RIF5 F10 | TTCTTAAGTACACGCTGAATCG | 59573 - 59594d | 414 | III |
| RIF5 R13 | GTCACAGTAATTCCATCAATGC | 59986 - 59965d | |||
| G | IS431 P4 | CAGGTCTCTTCAGATCTACG | 49963 - 49982c | 381 | II |
| pUB110 R1 | GAGCCATAAACACCAATAGCC | 50343 - 50323c | |||
| H | IS431 P4 | CAGGTCTCTTCAGATCTACG | 29654 - 29673d | 303 | III |
| pT181 R1 | GAAGAATGGGGAAAGCTTCAC | 29976 - 29956d | |||
| mecA | MECA P4 | TCCAGATTACAACTTCACCAGG | 1190 - 1211e | 162 | Internal control |
| MECA P7 | CCACTTCATATCTTGTAACG | 1351 - 1332e |
aLoci G and H were included to distinguish variants IA from I and IIIA from III, respectively.
bRelative to accession No. AB033763, SCCmec type I
cRelative to accession No. D86934, SCCmec type II
dRelative to accession No. AB037671, SCCmec type III
eRelative to accession No. Y00688, mecA gene
3.4. Statistical Analysis
The data were analyzed using Fisher’s exact test with SPSS, version 16 (SPSS Inc., U.S.). The level of significance was considered as 0.05.
4. Results
In total, the presence of MRSA in 262 samples was tested. Of the 148 samples from Kashani hospital, 76 (51%) were from males. Of the 114 samples from Hajar hospital, 48 (43%) were from males. Fisher’s exact test indicated no significant association between gender and the colonization rate of the MRSA isolates (P = 0.218). The mean age of the participants from Hajar and Kashani hospitals was 31.5 ± 20 and 32 ± 12 (range: 21 - 51 and 21 - 45) years, respectively. In addition, Fisher’s exact test indicated no significant association between age and the colonization rate of the MRSA isolates (P = 0.658). The proportion, age, and gender of the S. aureus-positive participants are shown in Table 2.
| Variable | Gender | Age | ||
|---|---|---|---|---|
| Group | Female | Male | ≤ 50 | > 50 |
| Frequency | 18 | 30 | 42 | 6 |
| % | 37.5 | 62.5 | 87.5 | 12.5 |
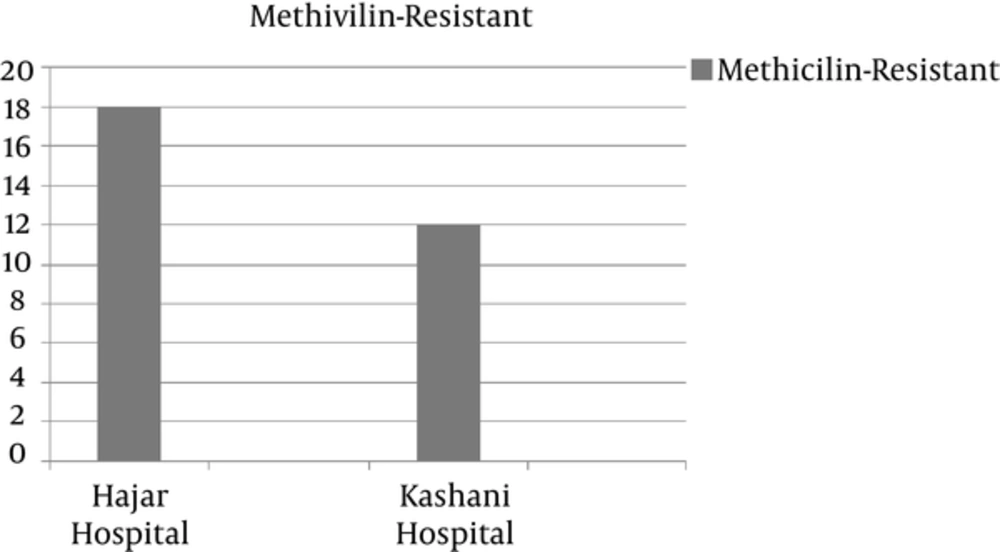
Nineteen of the 148 (12.8%) isolates from Kashani hospital were identified as S. aureus, of which 12 (63% [8% of total isolates]) were MRSA. In Hajar hospital, 29 of the 114 isolates were identified as S. aureus, of which 18 (62% [15.7% of total isolates]) were MRSA (Figure 1). Based on a coagulase-positive test, S. aureus was detected in 48 samples (18%). In the oxacillin disk-diffusion test of the methicillin resistance of the coagulase-positive isolates, 30 of the 48 S. aureus isolates were MRSA.
To determine the prevalence of MRSA carriers among staff with different occupations in the hospitals, the participants were divided into three groups (physicians, nurses, and nontreatment). The latter consisted of service personnel, kitchen staff, office staff (secretaries and administrators), and guardians. In Hajar hospital, nurses (50%) accounted for the highest proportion of carriers, and nontreatment staff accounted for the lowest proportion (22%). In Kashani hospital, nurses also comprised the highest proportion (41%) of carriers, and nontreatment staff comprised the lowest proportion (8%).
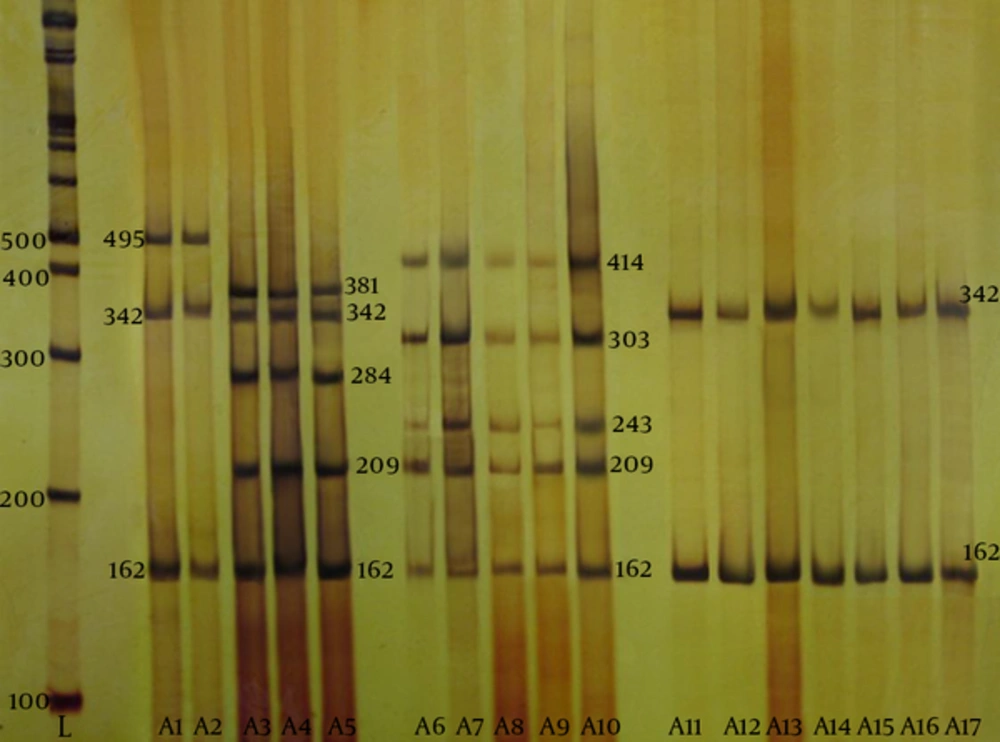
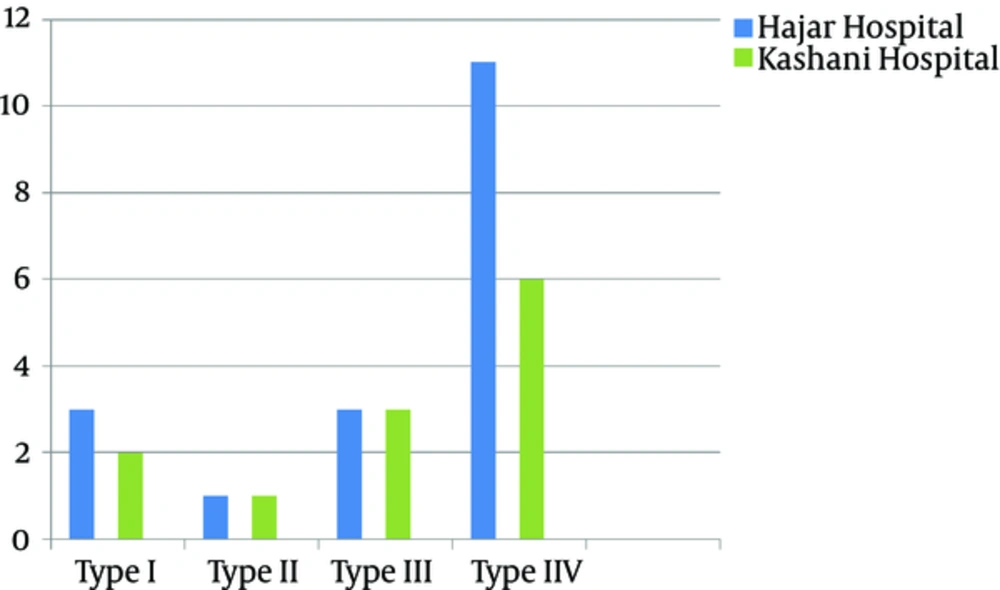
According to the antibiotic susceptibility analysis, the highest resistance obtained was to rifampin (33%), and all the isolates were susceptible to quinupristin-dalfopristin vancomycin, and linezolid. Table 3 summarizes the antibiotic susceptibility pattern of the MRSA isolates. The mecA gene was detected in all the MRSA isolates by the PCR. The results of SCCmec typing based on the multiplex PCR method showed that of 30 tested isolates, five (16.7%) were type I, two (6.7%) were type II, six (20%) were type III, and seventeen (56.6%) were type IV MRSA (Figure 2). Figure 3 shows the SCCmec types of methicillin-resistant isolates detected in the two hospitals.
| Antibiotic | Susceptible, No. (%) | Intermediate, No. (%) | Resistant, No. (%) |
|---|---|---|---|
| Bacitracin, 10 µg | 24 (80) | 0 | 6 (20) |
| Gentamycin, 10 µg | 26 (86.7) | 0 | 4 (13.3) |
| Novobiocin, 30 µg | 25 (83) | 1 (3) | 4 (13.3) |
| Ticoplanin, 30 µg | 18 (60) | 5 (17) | 7 (23) |
| Linezolid, 30 µg | 30 (100) | 0 | 0 |
| Quinupristin-dalfopristin, 15 µg | 30 (100) | 0 | 0 |
| Tigecycline, 15 µg | 17 (56.7) | 4 (13.3) | 9 (30) |
| Rifampin, 5 µg | 19 (63.4) | 1 (3.3) | 10 (33.3) |
| Vancomycin, 30 µg | 30 (100) | 0 | 0 |
From left to right, first lane, DNA marker; lane A1, standard SCCmec type 1; lane A2, SCCmec type 1; lane A3, standard SCCmec type 2; lanes A4 and A5, SCCmec type 2; lane A6, standard SCCmec type 3; lanes A7 to A10, SCCmec type 3; lane A11, standard SCCmec type IV; lanes A12 to A17, SCCmec type IV.
5. Discussion
The present study was conducted to determine the frequency and molecular types of MRSA isolated from nasal carriers in teaching hospitals in Shahrekord. In this study, 48 of 262 (18%) samples were identified as S. aureus, of which 30 (11.45%) were MRSA. In different studies of nasal samples obtained from healthcare staff, the prevalence of MRSA was reported to be 6.2%, 12.7%, 13.95%, 14.3%, 6.7%, and 18% in France, Ethiopia, Pakistan, India, and Saudi Arabia, respectively (27-32). Various prevalence rates (5.3 - 53.8%) of MRSA have been reported for Staphylococcus species isolated from individuals in different regions of Iran and from individuals in different occupations (33, 34). The different prevalence rates of this pathogen can be attributed to dissimilarities in healthcare policies (pattern of medication use), sample collection, nosocomial infection control, and the performance, education, and adherence of healthcare staff to hygiene-related recommendations, all of which potentially contribute to the distribution of resistant strains, including MRSA (30, 35-39).
In the present study, the colonization rate of MRSA isolates was not associated with age or gender. A previous study of the prevalence and antibiotic susceptibility pattern of nasal samples obtained from hospital staff and analyzed using the PCR method also found no significant difference in the carriage of this pathogen according to age and gender (33). However, Diawara et al. reported that age was significantly associated with being a carrier of this pathogen (40). Gebreyesus et al. demonstrated that women were more frequent carriers of the pathogen than men and that this finding was statistically significant (41). Several factors, such as the methodology of the study and occupational setting, may have contributed to these associations.
Nurses comprised 50% of the MRSA carriers in the present study. In other studies, nurses were also reported to be the most frequent carriers of MRSA (30, 32, 41, 42). The fact that nurses have more frequent contact with patients than other healthcare staff do and that they provide care to patients in different wards throughout the hospital likely explains this finding. As a result, nurses may have a greater risk than other staff of acquiring community-acquired MRSA. Therefore, the higher rate of MRSA acquisition among the nurses in the present study is not surprising.
Among methicillin-resistant isolates, the highest resistance was to rifampin, and all the isolates were susceptible to quinipristin-dalfupristine, vancomycin, and linzolid. In a previous study, 64.1% of MRSA isolates were resistant to amikacin, and 76.92%, 51.28%, 87.18%, 71.8%, 10.26%, 5.13%, 89.74%, and 61.54% were resistant to ceftriaxone, ciprofloxacin, erythromycin, gentamicin, mupirocin, rifampin, tetracycline, and tobramycin, respectively (43). In the same study, all the MRSA and methicillin-susceptible S. aureus isolates were susceptible to fusidic acid, linezolid, teicoplanin, tigecycline, and vancomycin (43). In another study, MRSA isolates also appeared to exhibit high sensitivity to vancomycin, teicoplanin, and trimethoprim/sulfamethoxazole (44). Various factors, such as age, duration of treatment, and geographical region, have been shown to contribute to MRSA drug resistance (36, 45, 46). The results of the SCCmec typing in the present study demonstrated that 16.7% of MRSA isolates were type I, 6.7% were type II, 20% were type III, and 56.6% were type IV.
In a study of specimens obtained in a hospital setting, a PCR analysis revealed that SCCmec type I was the most frequent type (58.9%), followed by SCCmec type II (19.9%), type III (11.0%), and type IV (8.2%) (35). In another study, most isolates were SCCmec types II and IV (47). In a study of isolates obtained from healthcare staff, approximately half the specimens were types IVa and V, and no specimens were type I (48).
The prevalence of MRSA among healthcare staff in the two hospitals in southwestern Iran varied, and the estimated prevalence was lower than average. The findings indicated that the MRSA isolates showed the highest resistance to rifampin and that all the isolates were susceptible to quinipristin-dalfupristine, vancomycin, and linzolid. SCCmec type IV was the most prevalent MRSA isolate. Given the growth in the resistance of these bacteria to antibiotics, including methicillin, implementing plans to detect, control, and restrict carriers of MRSA, particularly healthcare staff who are in direct contact with patients, is vital to prevent the transmission of MRSA isolates to hospitalized patients.