1. Background
Small noncoding RNAs (sRNAs) play multifunctional roles in bacterial gene expression and are recognized as key regulators in bacteria. Generally, the length of RNA regulators ranges from approximately 50 to 500 nucleotides, and they lack expressed open reading frames (1, 2). Transencoded sRNAs are the most extensively studied and best characterized sRNAs, which can control bacterial gene expression via multiple molecular strategies to regulate the expression of gene targets (eg, direct binding with complementary sequences in the target mRNA molecules) (3, 4). The activity of sRNA in accordance with its specific target has both positive and negative effects on gene expression.
Brucella melitensis is a Gram-negative, facultative, intracellular, pathogenic α-proteobacterium, which causes abortion and infertility in a wide range of animals, including sheep, goats, cattle, and pigs, and induces debilitating febrile diseases in humans (5, 6). Alpha-proteobacteria include animal pathogens (Brucella and Bartonella spp.), plant pathogens (Agrobacterium spp.), and plant symbionts (Sinorhizobium spp.). Although these pathogens infect a variety of hosts, they show similar mechanisms and common genes (7, 8). Additionally, α-proteobacteria share common features for proper association within the host. For instance, the importance of Hfq directs attention towards the importance of sRNA AbcR, although biological functions are not well-defined (9).
The AbcR molecules are among the most recently identified rhizobial sRNAs in sRNA research. They have been experimentally characterized in α-proteobacteria, including B. abortus (10), and the plant pathogen, A. tumefaciens (11). Caswell and colleagues identified a novel AbcR sRNA, which regulates the expression of ATP-binding cassette (ABC) transport systems, encoding genes in closely related bacteria, ie, B. abortus and A. tumefaciens. Additionally, they highlighted the role of AbcR sRNA in the virulence of B. abortus (10).
Although AbcR1 and AbcR2 sRNAs have similar nucleotide conformations in α-proteobacteria, they widely differ in regulatory functions; for instance, AbcR1 regulates gene expression, while AbcR2 does not play such a role in A. tumefaciens (11). Likewise, AbcR sRNA is encoded in the intragenic region of Sinorhizobium meliloti and plays a vital role in regulating the gene expression of branched-chain amino acid binding protein, LivK, by AbcR1, not AbcR2 (12).
Transcriptional regulators are important study topics at the molecular level. The family of transcriptional regulators continuously increases in size. With more than 800 members identified, it is known as the largest family of prokaryotic DNA-binding proteins (13, 14). It is involved in activities, including virulence, metabolism, motility, oxidative stress responses, and quorum sensing (15-18). The Two-component regulatory system showed that BvrR/BvrS, which is comprised of sensor and response regulator components, is responsible for signal transduction, environmental sensing, adaptive responses, and intracellular survival. It is also among the most effective Two-component regulatory systems of Brucella (19, 20).
2. Objectives
The present study aimed to characterize sRNA AbcR1 interaction with B. melitensis target mRNAs. We constructed a novel shuttle expression plasmid for gene expression, enabling stable replication in B. melitensis. In this study, we showed that sRNA AbcR1 interacts with target mRNAs to regulate gene expression, and transcription regulator and Two-component response regulators are direct targets of AbcR1.
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
Brucella melitensis 16 M strain was used in the current study, and its derivatives were normally grown in tryptic soy broth (TSB) at 37°C in a biosafety level-3 laboratory, as described in the literature (21). Moreover, pXG superfolder GFP plasmid series were provided by Professor Jorg Vogel (University of Würzburg) for cloning the coexpression of sRNA AbcR1 and target GFP fusion plasmids. Escherichia coli strain Top10 (Invitrogen) and B. melitensis 16 M competent cells were used in all the experiments of GFP fusion and sRNA AbcR1 expression. Antibiotics were added to the medium when appropriate at the following concentrations: 20 μg/mL of chloramphenicol, 50 μg/mL of kanamycin, 100 μg/mL of ampicillin, and 50 μg/mL of gentamicin.
3.2. 5’ RACE System
The transcriptional start site was detected with 5’ end rapid amplification of complementary DNA ends (RACE) as reported in the literature (22), using modifications of Takara Biochemicals, as suggested by the manufacturer.
3.3. Construction of Target and AbcR1 Plasmids
An E. coli-based Two-plasmid system was used for studying sRNA gene expression, established by Urban and Vogel to confirm the regulation of B. melitensis target mRNAs with AbcR1 (23). The B. melitensis target mRNA-encoding genes and sRNA AbcR1 were amplified via polymerase chain reaction (PCR) with primer pairs (BM target For/Rev or sRNA AbcR1 For/Rev), using genomic DNA (as a template extracted from B. melitensis 16 M) and Platinum® Pfx DNA polymerase (Invitrogen); all primer pairs are enlisted in the supplementary file, Appendix 1.
The amplified DNA fragments were digested with Nsi I and Nhe I and ligated with T4 DNA ligase. Then, they were cloned into the pZE12 derivative after treatment of polynucleotide kinase, as reported in the literature (23). The target genes included BMEI0513, BMEI1615, BMEII1022, BMEI0387, and BMEII0390 encoding transcriptional regulators, as well as BMEI1751 and BMEII0853 encoding Two-component response regulators.
To construct the target mRNA plasmid, pXG10-SF and PCR-amplified target DNA fragments were digested with Nsi I and Nhe I and ligated with T4 DNA ligase (23). Moreover, the PCR-amplified pJV300 and sRNA AbcR1 fragments were digested with Nsi I and Nhe I and ligated with T4 DNA ligase. Successfully cloned constructs were confirmed by PCR and sequencing. Target GFP plasmid with/without AbcR1 expression plasmid was cotransformed in E. coli Top 10 cells (Invitrogen).
3.4. Construction of AbcR1 and Target mRNA Mutants
The bioinformatic prediction revealed that AbcR1 has a conserved region in all the selected target mRNAs. We constructed an AbcR1 mutant, using ΔAbcR-For/Rev primers and PCR-amplified fragments with DNA polymerase, using genomic DNA from B. melitensis 16 M; consequently, CCC was removed from the seed region. The gel-purified, digested fragment was ligated with pJV300 backbone, using T4 DNA ligase. Successfully grown plasmid was obtained in Top 10 competent cells, and the final authenticity of cloned pJV300-ΔAbcR1 construct was confirmed via PCR and DNA sequencing.
The point mutation was introduced in target mRNAs by changing the nucleotide base pair in the seed region of target mRNA (designated M). The point mutation was introduced as the following nucleotides: GAA with CUU in BMEI0513, AAC with UUG in BMEI1615, GGA with CCU in BMEII1022, AGA with UCU in BMEI0387, GAA with CUU in BMEII0390, AGA with UCU in BMEI1751, and AGA with UCU in BMEII0853 mRNA. Using the subsequent oligonucleotide pairs (supplementary file, Appendix 1), PCR was applied to amplify fragments with DNA polymerase with genomic DNA from B. melitensis 16 M. The gel-purified, digested fragment was ligated with pJV300 backbone, using T4 DNA ligase. The authenticity of each mutant plasmid was confirmed by PCR and DNA sequencing.
3.5. Construction of pZF17-08 Plasmid
Initially, pJV300 linear fragment was PCR-amplified, using pJV300 as the template. PmlI restriction site was introduced, resulting in the construct pZK001. The DNA sequence, containing pBBR1MCS-5 replicon and gentamicin-resistant gene, was PCR-amplified using pBBR1MCS-5 as the template. The linear fragments of pZK001 and pBBR1MCS-5 were combined via in-fusion cloning, producing the construct pZK41. Replication C and partition B genes were PCR-amplified from Ochrobactrum anthropi using primer repC F/R; all primer pairs are listed in the supplementary file, Appendix 1. The linear fragments of O. anthropi and pZK41 were combined using in-fusion cloning, producing the construct O. anthropi pZF17-08 plasmid.
3.6. Determination of Plasmid Copy Number
Real-time quantitative PCR (RT-qPCR) was used to determine the relative copy number of O. anthropi pZF17-08 plasmids. Three fragments were PCR-amplified using B. melitensis 16M gDNA, pBBR1MCS-5 plasmid, and pZF17-08 O. anthropi plasmid. Then, they were cloned into pMD18-T, using in-fusion cloning to construct a reference plasmid. The plasmid copy number was determined using the following formula:
Nrelative = (1 + E) −ΔCT
Where E represents the total amplification efficiency of the plasmid and reference, and −ΔCT is the difference between the threshold cycle number of the reference and plasmid (24). The used primers are listed in the supplementary file, Appendix 1. The samples were amplified accordingly for each 25-μL volume reaction, containing 5 pmol of forward and reverse primers, 1 μL of cDNA sample, and 12.5 μL of 2X SYBR green I master mix (Takara Biochemicals). The thermocycler reaction conditions were adjusted at 95°C for 3 minutes, followed by 45 cycles at 95°C for 20 seconds, 60°C for 60 seconds, and 95°C for 40 seconds; all the samples were assayed in 3 individual repeats.
3.7. Fluorescence Analysis in Agar Plates
Top 10 cells, expressing cotransformed plasmid GFP fusions and AbcR1, were streaked on Luria-Bertani (LB) agar plates. The plates were prepared as the medium with ampicillin and chloramphenicol resistance and streaked for subsequent colonies. After overnight growth, the green fluorescence images of the colonies were acquired in a stereomicroscopic image analyzer, using a CCD camera with an excitation/emission wavelength of 485/525 nm.
3.8. Fluorescence Measurement in Liquid Cultures
The established Two-plasmid superfolder GFP reporter system was used for rapid validation of the predicted interaction of sRNA AbcR1 with target mRNAs (23, 25). The GFP fluorescence of cotransformed plasmids was accessed in colonies grown on LB agar plates or cells obtained in liquid LB culture. To determine the whole-cell fluorescence, 96-well microtiter plates were used. Single colonies (3 independent replicates) of E. coli Top10 cells, harboring both target GFP fusion plasmid and sRNA AbcR1 expression plasmid, were cultured into a fresh 20-mL LB medium; the overnight grown cultures were shaken at 220 rpm at 37°C, and cell density was measured at OD600.
At the designated cell density, 150 μL of 3 independent aliquots from each grown culture was transferred into a microtiter plate and overlaid with mineral oil to avoid evaporation. Fluorescence was determined at 37°C (optical excitation filter, 480/31 nm; emission filter, 520/10 nm, 0.2 seconds; CW lamp energy, 21673; measurement height, 8.0 mm) in a Victor 3 machine (1420 Multilabel Counter, Perkin Elmer).
3.9. Western Blot Analysis
The cotransformed plasmids were grown overnight. The samples were spun at 12 000 rpm for 3 minutes at OD600. The supernatant was discarded, and the pellet was dissolved in 1 mL of PBS; also, 10 μL of × 100 concentrated cocktail was added to prevent inactivation of protein denaturation. The suspension was then clarified via ultrasonic disintegration at 5-second intervals until it looked completely clear. Following that, 5X SDS loading buffer was added and heated at 95°C for 10 minutes. The samples were placed on ice for cooling for about 2 minutes, and the aliquots were carefully handled to avoid repeated freezing and thawing.
Protein fractions harboring the target GFP fusion and sRNA AbcR1 fusion were separated by running on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were transferred at 80 V for 1 hour via BioRad electrophoresis on PVDF membrane (PerkinElmer) in the presence of a transmembrane buffer. The membranes were rinsed in 10% tris-buffered saline with Tween 20 (TBST), blocked with bovine serum albumin (BSA) for 4 hours, and washed 3 times with 10% TBST buffer.
The membranes were incubated in an anti-EGFP monoclonal antibody, mixed in 3% BSA at a concentration of 1:50000, and kept overnight at room temperature. After 10 minutes of washing in 10% TBST, the membranes were incubated into HRP-conjugated goat anti-mouse IgG antibody and mixed with 3% BSA at a concentration of 1:5000 at room temperature for 4 hours. GroEL was detected as the loading control with anti-GroEL polyclonal antibody (Sigma). The final images of the membranes were acquired using a Western lightning reagent (BioRad), and the signals were detected with a Fuji LAS-3000 CCD camera.
3.10. RNA Isolation From Cotransformed Plasmids
Trizol reagent (Invitrogen) was used to extract total RNA from GFP target fusion plasmid and AbcR1 expression plasmid, according to the manufacturer's protocol (Takara Biochemicals). The samples were grown overnight for RNA isolation to an OD600 of 1. To eliminate genomic DNA from cotransformed plasmids, RNA samples were treated with DNase I (Promega). Agarose gel electrophoresis and spectrophotometry (Nanodrop Technologies) were used to determine the quality and quantity of isolated RNA.
3.11. Quantitative RT-PCR
Expression of target GFP-fused plasmid and sRNA AbcR1 expression plasmid was measured via quantitative RT-PCR (qRT-PCR; Applied Biosystems); also, 16S rRNA was used as the reference gene to standardize the expression data for each target. Relative transcriptional level was determined using the delta-delta CT method, as previously described (26).
3.12. Biocomputational Predictions
Information about the target gene sequences of B. melitensis 16M was collected using Webtool (http://www.ncbi.nlm.nih.gov/genome/943). Moreover, target mRNA and AbcR1 interaction sites were predicted with intaRNA (27) programs.
3.13. Statistical Analysis
Paired student t test was used to analyze the data. To determine the significance of AbcR1 expression by qRT-PCR, Wilcoxon signed-rank test was applied. P-value below 0.05 was assumed to be statistically significant.
4. Results
4.1. Annotation and Prediction of sRNA AbcR1 in B. melitensis
Computational genome annotation was performed to extract the AbcR1 sequence from B. melitensis 16M chromosome (NC-003317.1 and NC-003318.1), based on genome annotation. Target site predictions were performed using the intaRNA Webtool to determine the regions of interaction between target mRNAs and AbcR1. The AbcR1 sequence was subjected to a BLAST search against B. melitensis 16 M chromosome in intaRNA. P-value less than 0.05 was considered significant for Brucella and excluded from further analysis.
Putative pseudogenes, origins of replication, and sequences encoding rRNAs and/or tRNAs were also excluded. Target genes were predicted based on 80-nt upstream and 20-nt downstream of mRNA with minimum length of 7-nt seed region and interaction region of < 20 nt (sRNA-mRNA structural accessibility and conservation assumed as true). Based on these criteria, a total of 7 candidate target mRNA genes were selected from the transcriptional regulator family (BMEI0513, BMEI1615, BMEII1022, BMEI0387, and BMEII0390) and Two-component response regulators (BMEI1751 and BMEII0853). Furthermore, attention was paid to avoid shift mutations; the sequences did not encode small peptides. The predicted target mRNAs, location on chromosome, gene orientation in B. melitensis 16 M, gene length, binding region with AbcR1, and putative function are shown in Table 1.
| No. | Predicted Target mRNA | Chromosome | Gene Orientation | Length | Binding Region With AbcR1 | Function |
|---|---|---|---|---|---|---|
| 1 | BMEI0513 | NC003317 / AE008917 | →→← | 533836 - 534741 | 40 - 48 | lysR family of transcriptional regulators |
| 2 | BMEI1615 | NC003317 / AE008917 | ←←← | 1661685 - 1662692 | 14 - 21 | araC family of transcriptional regulators |
| 3 | BMEII1022 | NC003318 / AE008918 | ←→→ | 1061455 - 1062309 | 17 - 25 | transcriptional regulators |
| 4 | BMEI0387 | NC003317 / AE008917 | ←→→ | 396715 - 397536 | 24 - 35 | iclR family of transcriptional regulators |
| 5 | BMEII0390 | NC003318 / AE008918 | ←→→ | 409693 - 410625 | 39 - 49 | lysR family of transcriptional regulators |
| 6 | BMEI1751 | NC003317 / AE008917 | ←←→ | 1793505 - 1794197 | 53 - 67 | two-component response regulator |
| 7 | BMEII0853 | NC003318 / AE008918 | ←←→ | 889705 - 890472 | 60 - 71 | two-component response regulator |
4.2. Experimental Validation and Regulation of sRNA AbcR1 Expression in B. melitensis Targets
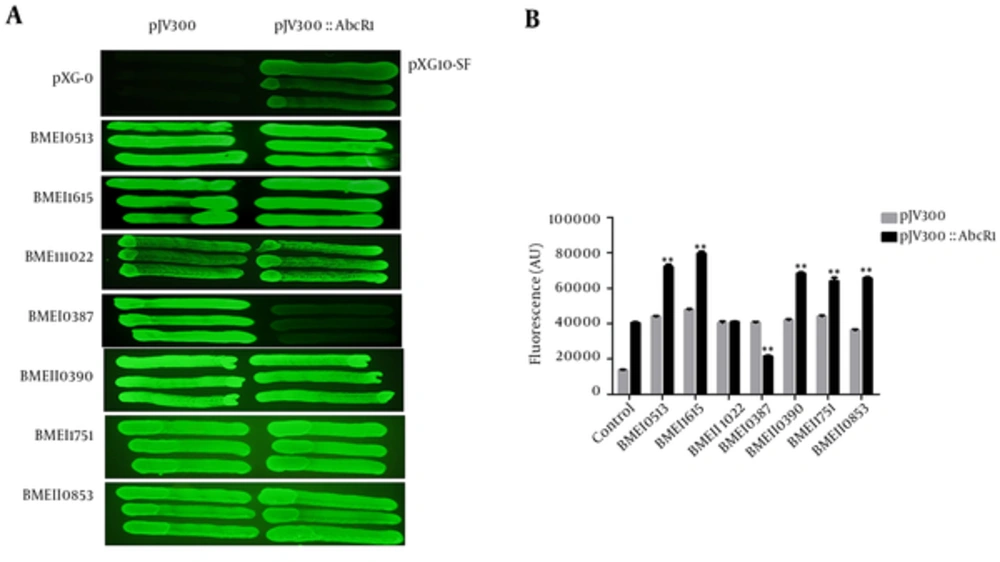
The Two-plasmid superfolder GFP reporter system was used to determine the expression of sRNA AbcR1 regulation and potential interactions with B. melitensis target mRNAs. We constructed GFP target fusion plasmids and AbcR1 expression plasmids to confirm fluorescence variations. The preliminary fluorescence was determined by streaking colonies on standard LB agar plates, which revealed significant variations among GFP target fusion plasmids with AbcR1 expression plasmid, compared to cotransformed plasmids harboring no AbcR1 (Figure 1A).
(A) Representative fluorescence image of B. melitensis target mRNA expression on the LB agar plates. The target mRNAs are cotransformed with pJV300 plasmid (without AbcR1) and pJV300-AbcR1 (AbcR1 expression plasmid). Images are acquired in the fluorescence mode at an excitation/emission wavelength of 485/525 nm. (B) Comparison of fluorescence values of overnight grown cotransformed plasmids in fresh LB medium, containing ampicillin and chloramphenicol antibiotics; fluorescence is determined at a cell density (OD600) of 1. In the control column, the values indicate pXG-0 and pXG10-SF plasmids without the target and AbcR1. The fluorescence of each target is calculated in arbitrary units (AU) as the mean fluorescence of 3 independently grown cultures.
We aimed to determine the GFP activities of a broad set of GFP target fusion plasmids, as well as AbcR1 expression plasmids in liquid cultures. The cultures were incubated with aeration at 37°C/220 rpm, and cell density was determined, followed by measuring OD600 of 1. The positive control plasmid, pXG10-SF-pJV300, showed higher GFP fluorescence than the pXG-0-pJV300 plasmid (negative control) and expressed the working system. Target mRNAs, ie, BMEI0513, BMEI1615, and BMEII0390, exhibited higher fluorescence with AbcR1 in comparison with non-AbcR1 expression plasmid.
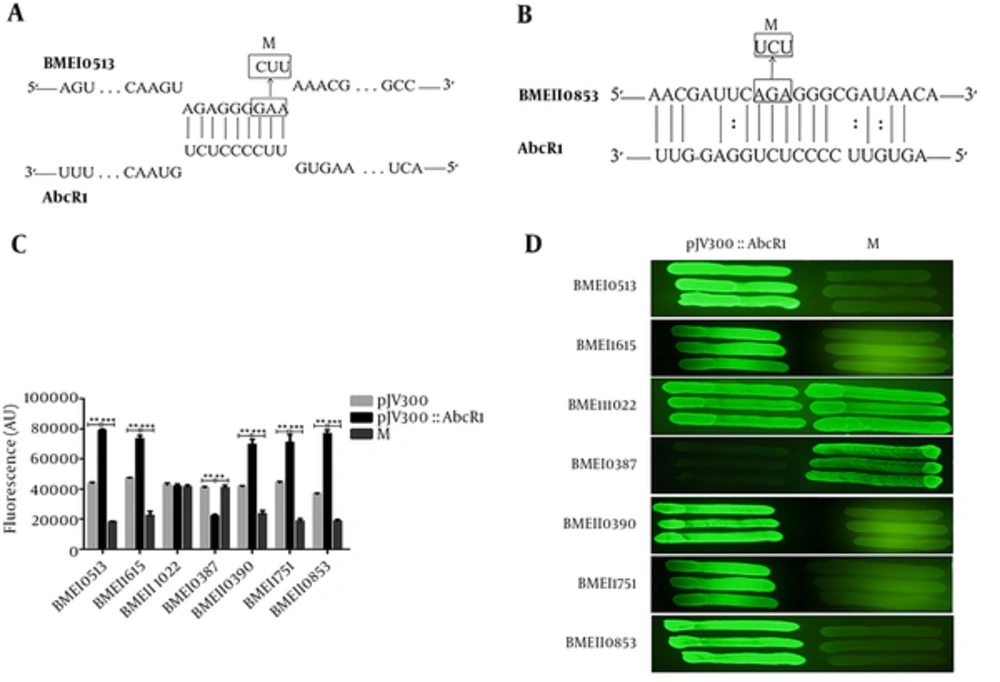
The fluorescence of BMEII1022 GFP target fusion with AbcR1 was intermediate and similar to fluorescence without AbcR1 expression plasmid. The fluorescence of BMEI0387 GFP target fusion plasmid with AbcR1 was lower than the plasmid without sRNA AbcR1, showing the downregulation of target mRNA with sRNA AbcR1 interaction. The Two-component response regulators, BMEI1751 and BMEII0853 GFP target fusion plasmids, exhibited higher fluorescence with AbcR1, compared to plasmids without sRNA AbcR1 (Figure 1 B). At an OD600 of 1, targets without AbcR1 displayed a consistent fluorescence of 35 000 to 43 000 arbitrary units (AU), which is nearly twice the fluorescence of AbcR1 expression plasmids; therefore, AbcR1 positively regulated these targets.
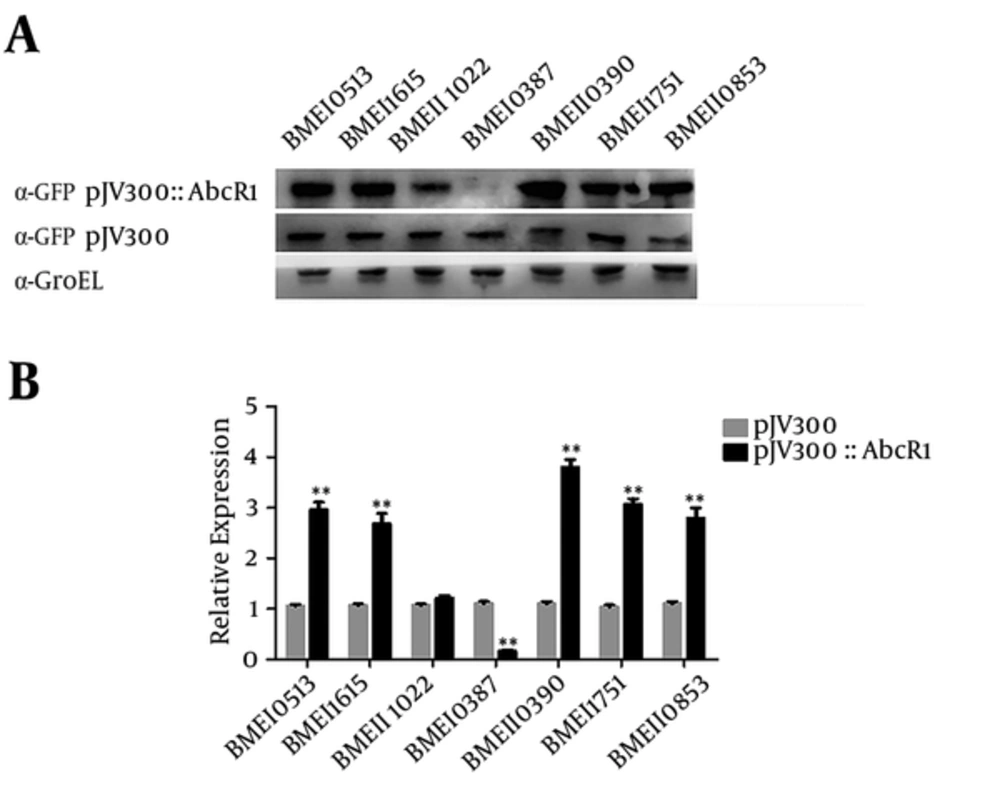
To determine sRNA AbcR1 regulation more accurately, we measured the fusion protein expression via Western blot analysis from cotransformed plasmids as pXG10-SF/BM target with pJV300 and pJV300-AbcR1. In 5 target cases, GFP fluorescence, as well as protein level, was higher with AbcR1, confirming that AbcR1 positively regulates mRNA targets. However, in BMEII1022, target protein expression was similar with and without AbcR1, showing its negligible effect. In contrast, BMEI0387 had lower fluorescence in the presence of AbcR1, and protein was barely detectable at mRNA level (Figure 2 A). These results confirm the high specificity and strong regulatory interactions between target mRNAs and AbcR1 sRNA.
(A) Detection of fusion protein expression of cotransformed plasmids by Western blot analysis. Samples are taken from cotransformed plasmid liquid cultures at an OD600 of 1 and subjected to Western blot analysis with anti-GFP monoclonal antibody, where loading control is performed with anti-GroEL polyclonal antibody. (B) RT-PCR verification of AbcR1 regulation by extracting total RNA from the cotransformed plasmids of 3 individual replicates from each sample culture, normalized with 16 S rRNA. The relative transcription of the predicted target mRNA regulation is measured in the presence and absence of AbcR1.
We further validated the predicted target mRNAs via qRT-PCR and normalized them with 16 S rRNA. Total RNA was isolated from the cotransformed plasmids of 3 individual replicates from each sample culture. To verify AbcR1 regulation interaction, target mRNA expression was compared with AbcR1 via qRT-PCR, which showed BMEI0513, BMEI1615, BMEII1022, BMEI1751, and BMEII0853 target mRNAs to be strongly upregulated in the presence of AbcR1; therefore, the results of GFP florescence and Western blot analysis were supported (Figure 2 B). BMEII1022 expression was higher with AbcR1. However, BMEI0387 showed downregulation with AbcR1, compared to the target without AbcR1, although the signal was stronger than that indicated by fluorescence and Western blot analyses.
4.3. Conserved Seed Regions in sRNA AbcR1 Regulate Multiple Targets of B. melitensis
Our previous findings indicated that AbcR1 directly interacts at the mRNA level of target genes to facilitate gene expression. Although AbcR1 contains binding regions specific to target mRNAs, conserved nucleotides were found in the interaction region of AbcR1 in all selected target mRNAs (Figure 3 A). We showed that the conserved nucleotide CCC in the seed region of AbcR1 coincided with the binding region of all selected target mRNAs. To investigate the role of AbcR1 interaction with target mRNA, we constructed an AbcR1 mutant and cloned it in pJV300 plasmids. Fluorescence was determined after cotransformation with GFP fusion plasmids, containing the target mRNA.
(A) AbcR1 harbors highly conserved nucleotides, which coincide in all the selected target mRNAs in the seed interaction region. AbcR1 sequence is shown against B. melitensis target mRNAs, where the seed region of AbcR1 is indicated in red color showing conserved nucleotides. (B) The fluorescence of overnight grown target plasmids, cotransformed with pJV300, pJV300-AbcR1, and pJV300-ΔAbcR1, is compared. The fluorescence of each plasmid is calculated in arbitrary units (AU).
Seven selected targets (BMEI0513, BMEI1615, BMEII1022, BMEI0387, BMEII0390, BMEI1751, and BMEII0853) showed significant up/downregulation with mutant AbcR1 (Figure 3 B); therefore, conserved nucleotides in the seed region of AbcR1 play an essential role in gene regulation. The target BMEI0387 showed downregulation in our initial experiment, while the fluorescence of the target infused with sRNA ΔAbcR1 was equal to that of the control plasmid, providing further evidence that AbcR1 regulates gene expression.
In contrast to the conserved region of AbcR1, regulation of target mRNA fusion was dependent on the seed pairing of AbcR1. To confirm AbcR1 targeting of mRNAs via seed pairing, we used the intaRNA algorithm to predict AbcR1-mRNA interaction, querying seed pairing of these targets (Figure 4 A). The predicted targeting region of AbcR1 interacted with the region near the early coding DNA sequences (CDS) of the target; as in prototypical target regulation, AbcR1 seems to act at the level of translation initiation. To validate this prediction, we constructed point mutation via nucleotide change in the seed region of target mRNA (designated M).
The schematic representation of the interaction of AbcR1 with (A) BMEI0513 (transcriptional regulator) and (B) BMEII0853 (Two-component response regulator) mRNAs, as well as changes in the nucleotide base pair. The numbers indicate the relative direction of sRNA AbcR1, as well as BMEI0513 and BMEII0853 mRNAs. The arrows indicate nucleotide substitutions (inbox) introduced to BMEI0513 and BMEII0853 mRNAs. M indicates plasmids with nucleotide changes in BMEI0513 and BMEII0853. (C) The fluorescence of overnight grown cotransformed plasmids was calculated in the fresh LB medium, containing ampicillin and chloramphenicol at a cell density (OD600) of 1. (D)5 The fluorescence image of target mRNA expression on the LB agar plate is presented.
As predicted, this mutation mirrored the inability of AbcR1 to regulate the target, and fluorescence values of cotransformed plasmids were compared with and without AbcR1 in the microtiter plate (Figure 4 C), as well as LB agar plates (Figure 4 D). The target mRNAs were significantly up/downregulated after mutation. This finding supports the prediction that seed pairing of AbcR1 plays an essential role in target mRNA regulation and confirmed the direct AbcR1 regulation in the expression of genes encoding transcription regulators and Two-component response regulators in B. melitensis. The transcription regulator and Two-component response regulators were direct targets of AbcR1.
4.4. Characteristics of the Shuttle Plasmid and Generation of a Standard curve to Determine the Copy Number
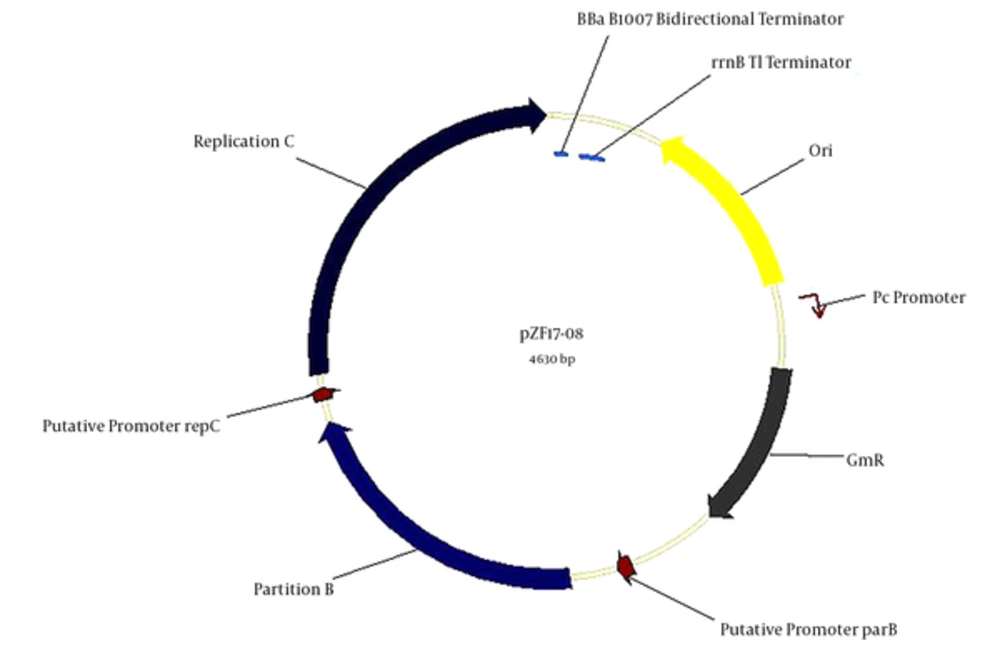
We constructed a novel shuttle plasmid to validate sRNA-mRNA interaction in Brucella. The genetic organization of plasmid pZF17-08 is shown in Figure 5. The detailed construction strategy has been described in the materials and methods section. This plasmid contained a terminator sequence (rrnBT1), a BBa B1007 bidirectional terminator, gentamicin as the selectable marker, and elements for replication such as replication initiator protein (repC) and replication-partition system (28). As this plasmid carries a gentamicin-resistant gene in Brucella, the construct pZF17-08 was electrotransformed into B. melitensis 16M competent cells, which was shown to trigger plasmid replication.
The plasmid melting curve analysis showed melting temperatures of 85.62 ± 0.18°C, 85.29 ± 0.22°C, and 85.89 ± 0.15°C for the templates of pZF17-08, pBBR1MCS-5, and B. melitensis, respectively. The slopes of the standard curves were -3.4154, -3.4112, and -3.4278, while the r-square values (R2) were 0.9999, 0.9997, and 0.9985, respectively. Moreover, the amplification efficiency (E value) was 96.242, 96.405, and 95.983 for pZF17-08, pBBR1MCS-5, and B. melitensis, respectively. This measurement was also valid, as the amplification efficiencies of pZF17-08, pBBR1MCS-5, and B. melitensis were almost identical (Figure 6). The copy number of pZF17-08 plasmid was calculated as 1.
4.5. Validation of sRNA AbcR1 Regulation in B. melitensis 16 M Competent Cells
To determine the target gene expression more accurately, we transformed the target mRNA plasmid in B. melitensis 16 M competent cells, which enabled stable replication in B. melitensis. The target mRNAs were individually cloned into pZF17-08 plasmids. We constructed point mutations by changing the nucleotide base pair in the seed region of target mRNA (designated M); they were cloned into pZF17-08 plasmids to construct the target mutant plasmid. We transformed the plasmids in B. melitensis 16 M competent cells to observe the regulatory effects. The target mRNAs showed significant up/downregulation, and the fluorescence values were determined in the microtiter plates, as shown in Figure 7 A.
The target mRNAs, BMEI0513, BMEI1615, BMEII0390, BMEI1751, and BMEII0853, exhibited higher fluorescence, compared to mutant plasmids. The fluorescence of BMEII1022 was intermediate, and fluorescence of BMEI0387 plasmid was lower than that of the mutant plasmid, thus showing the downregulation of target mRNA. Furthermore, we confirmed this interaction by Western blot analysis, as shown in Figure 7 B. We further validated target mRNA expression via qRT-PCR and normalized it with 16S rRNA. Total RNA was isolated from the transformed plasmids of 3 individual replicates from each sample culture. The target showed significant up/downregulation, compared to the mutant plasmids (Figure 7 C). These results confirm the high specificity and regulatory interaction between target mRNAs and sRNA AbcR1.
5. Discussion
Brucella species are facultative intracellular pathogens, which naturally infect a wide range of host animals, such as sheep, goats, and cattle (5). Brucella employs multiple strategies through posttranscriptional regulation of gene expression to cope with changing environments (29). Moreover, sRNAs can regulate gene expression and/or allow for rapid alterations to encounter stimuli and stress (30). While more than 200 sRNAs have been experimentally validated in bacteria, such as Salmonella enterica and E. coli, only 5 sRNAs from Brucella have been studied so far (31, 32). In the present study, we identified and validated mRNA-AbcR sRNA interactions with B. melitensis target mRNAs (which belong to the transcription regulator family), as well as Two-component response regulators.
The pZF17-08 plasmid carries the terminator sequence (rrnBT1), BBa B1007 (29). Gentamicin-resistant gene is shown in grey, and the arrow indicates the direction. The replication C and partition B are shown in blue.
The r-square, slope, and intersection values of (A) pZF17-08, (6) pBBR1MCS-5, and (E) B. melitensis of qPCR tests. The melt curves of (B) pZF17-08, (D), pBBR1MCS-5, and (F) B. melitensis are also shown in this figure.
(A) The fluorescence values of B. melitensis target mRNA expression in the microtiter plates, (B) detection of fusion protein expression of transformed plasmids by Western blot analysis, (C) RT-qPCR verification of AbcR1 regulation by extracting total RNA from the transformed plasmids of 3 individual replicates from each sample culture, normalized with 16S rRNA.
Using bioinformatics prediction tools, we selected 7 target candidate genes in B. melitensis and identified their interactions with AbcR1 sRNA. Using intaRNA, we predicted the interaction between target mRNA and sRNA AbcR1, based on the common features of transencoded RNAs. Similar studies have predicted and validated the interaction of MicF RNA with target mRNAs in Salmonella (33), as well as the interaction of GntR regulators with target mRNAs involved in intracellular survival of Brucella (34).
Transcriptional regulators and Two-component response regulators have been reported to play significant roles in the regulation of diverse biological processes, particularly in the virulence of B. melitensis (35). In the present study, the GFP reporter system was used to validate the interaction of target mRNAs with AbcR1 sRNA. Five out of 7 targets showed upregulation, while BMEI0387 was downregulated. However, fluorescence of BMEII1022 target GFP fusion was intermediate and similar to the fluorescence of AbcR1 expression plasmid.
The interaction of target mRNAs with AbcR1 was further confirmed by Western blot and qRT-PCR analyses. Similar studies have validated several ABC transporter mRNAs as AbcR1 targets in A. tumefaciens and B. abortus (10, 11), and sRNA AbcR1 has been reported as a master regulator in α-proteobacteria (36). Previous studies have reported the interactions of mRNAs from the global transcriptional regulator, leucine-responsive regulatory protein (Lrp), and YahO with MicF RNA (33, 37). Our findings were validated in a similar study of Salmonella, providing strong evidence of MicF-Lrp interaction (33).
The AbcR sRNA showed high conservation in α-proteobacteria. We identified an AbcR1-conserved region in all the predicted target genes. To further clarify the assumption that the conserved seed region was essential to the regulation of target mRNAs, we constructed a mutant of sRNA AbcR1; sRNA ΔAbcR1 affected the regulation of all predicted target mRNAs. All the selected targets showed significant up/downregulation with the mutant AbcR1 (Figure 3 B), showing the essential role of conserved nucleotides in the seed region of AbcR1 in gene regulation. This supports the finding that B. abortus VtlR transcription regulator target mRNAs (bab1-0310, bab1-0313, bab1-0314, and bab1-1794) are regulated by AbcR sRNAs and show higher regulatory gene expression with ΔAbcR sRNAs (38).
In addition, AbcR sRNAs were reported to control the expression of Lrp-encoded gene BAB1-1587 in B. abortus, and expression decreased with ΔAbcR sRNAs; this finding highlights the importance of AbcR sRNA in the regulation of metabolism and amino acid transport (10). In contrast to the conserved region of AbcR1 sRNA, the regulation of target mRNA fusion was dependent on the seed pairing of AbcR1 sRNA. We validated this prediction by changing the nucleotide base pair of target mRNAs. The target mRNAs were significantly up/downregulated after this mutation.
Similarly, a previous study showed that the interaction of GntR transcriptional regulator with BSR0602 sRNA was validated by introducing point mutations in sRNA and target mRNA in B. melitensis (34). Our findings were validated in a parallel study on E. coli where MicF-Lrp interaction (37), as well as MicF interaction with Lrp and YahO in Salmonella, was identified and validated (33). We constructed a novel shuttle plasmid to validate the sRNA-mRNA interaction in Brucella. We transformed the plasmids in B. melitensis 16 M competent cells to observe the regulatory effects. The target mRNAs showed significant up/downregulation, and the fluorescence values were observed in the microtiter plates, as shown in Figure 7.
6. Conclusion
Our findings suggest that sRNA AbcR1 has a conserved seed region, which interacts with the target mRNAs of transcriptional and Two-component regulators to control their gene expression. The conserved region of AbcR1 regulates multiple target mRNAs by interactions within the seed region. The conservation and regulatory interactions of sRNA AbcR1 with transcriptional regulator family and Two-component regulators highlight the role of sRNA AbcR1 in B. melitensis.