1. Background
One of the main agents of infectious diseases is methicillin-resistant Staphylococcus aureus (MRSA) (1). This pathogen causes a series of infections in soft tissues and skin, such as bacteremia, endocarditis, pneumonia, toxic shock syndrome, osteomyelitis, and septicemia (2). Methicillin-resistant S. aureus are bacteria responsible for several difficult-to-treat infections in humans and high rate mortality due to the process of natural selection. These infections are more difficult to treat with common standard antibiotics, so they are a potential risk for the human health (3, 4). These bacteria are the most frequent agents in the community and hospitals. Community-associated methicillin-resistant S. aureus (CA-MRSA) distribution has changed globally (5, 6).
Resistance to beta-lactam antibiotics in MRSA is caused by mecA, the gene encoding penicillin-binding protein (PBP2a) with the low affinity to beta-lactams, which was first discovered in 1960 (7). Methicillin-resistant Staphylococcus aureus could be prevented if penicillin-binding proteins are intact (8). The emergence of resistance to MRSA has developed the need for novel strategies to replace alternatives to traditional antibiotics (9). Nanotechnology is gaining much attention in the current century due to its capability to modify properties of metals into their nanoscale (10). Recently, researchers have identified nanotechnology as promising to the antimicrobial agent (11). Antimicrobial mechanisms of nanomaterials include disruption of cell membrane, production of reactive oxygen species, and inhibition of enzyme activity, DNA synthesis, and interruption of energy transduction (12).
One of the nanomaterials is metal-organic frameworks (MOFs), which are porous, hybrid materials containing metal ions linked by organic binding ligands. In addition, these nanostructures have demonstrated significant antibacterial activity and their properties make the MOFs promising candidates for medical applications (13). One of the metals used is the nanostructure copper, which is showing antimicrobial features. The major mechanisms of copper are cytoplasmic membrane damage or DNA damage with the production of reactive oxygen species (ROS) (14, 15). Consequently, the synergistic effect between the presence of copper and stable structures has been expected to mainly improve the antibacterial effect. Considering the increase in resistance of antibiotics of MRSA and useful properties of the nanostructure, we designed then synthesized the Cu-BPDCA-Ty nanostructures.
2. Objectives
The present study aimed at evaluating the antibacterial potential of Cu-BPDCA- Ty and its effect(s) on the expression of mecA gene of MRSA.
3. Methods
3.1. Microbial Strains
Bacterial strains were used in MRSA (ATCC 33591, mecA positive), MSSA (ATCC25923, mecA negative), and 5 clinical strains of MRSA. Clinical strains were obtained from the microbiology laboratory of the school of medicine of Yasuj University of Medical Sciences.
3.2. Phenotypic and Genotypic Characterization of MRSA
Conventional biochemical tests such as catalase, tube coagulase, fermentation of manitol salt agar (MSA), and DNase were used to identify clinical strain of S. aureus. Presence of S. aureus isolates were confirmed by amplifying the nucA gene. Cefoxitin containing disk was used for phenotypic identification of MRSA strains, according to the clinical and laboratory standards institute guideline (CLSI) (16). The mecA gene was amplified by PCR method for molecular confirmation of MRSA isolates, as described previously. Also, we tested susceptibility pattern of the isolates against. Primers were used at a concentration of 10 picomoles. The primers used for the detection and the amplification protocol are listed in Tables 1 and 2, respectively.
| Gene | The Initial Denaturation | Denaturation | Annealing of Primers | Extension | Final Extension | Cycle |
|---|---|---|---|---|---|---|
| mecA | 94°C (4 min) | 94°C (40 s) | 56°C (40 s) | 72°C (40 s) | 72°C (5 min) | 30 |
| nucA | 94°C (4 min) | 94°C (40 s) | 58°C (40 s) | 72°C (40 s) | 72°C (5 min) | 30 |
3.3. Synthesis of Cu-BPDCA -Ty
The Cu-BPDCA-Ty was prepared by ultrasound-assisted solvo thermal method as follows: Thymine (0.125 mmol) and biphenyl dicarboxylate acid (0.25 mmol) were mixed for 30 minutes in 100 mL of solvent with a volume ratio (70 DMF: 30 H2O). The ultrasonic bath was used for the mix. Then, nitric acid (1 mmol) was added for 30 minutes under ultrasonic waves. Following copper (II), nitrate (Cu (NO3)2) (0.375 mmol) was added to the solution for 30 minutes. Subsequently, the mixture was transferred into a Teflon lined autoclave, heated at 130°C for 24 hours, and then cooled to room temperature naturally. Finally, the precipitate was washed with H2O and ethanol and dried in an oven at 120°C (19).
3.4. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
The minimum inhibitory concentration (MIC) of nanostructure was determined by broth microdilution method using sterile 96-well microplates. Concentrations used ranged from 0.1 to 1 mg/mL. The final volume of the well was 100 μL. The volume required from the culture medium and the nanostructure was calculated using the following formula: C1V1 = C2V2. The serial dilution with sterile Mueller Hinton broth (MHB) was transferred into well of a microplate and inoculated with 5 μL of standardized (4 - 5 × 105 CFU/ mL) bacterial suspension. Moreover, the microplate was incubated aerobically at 37°C for 18 to 24 hours. Subsequently, the lowest concentration of the nanostructure prevented the growth of bacteria as MIC. The positive control contained the bacteria inoculated into MHB, without Cu-BPDCA-Ty, and the negative control was only media, without bacteria and Cu-BPDCA-Ty. Finally, turbidity was measured using a microplate reader (BiotecElx 800) at the wavelength of 630 nm. In addition, the minimum bactericidal concentration (MBC) was performed by subculturing 100 μL/mL of wells without turbidity on the blood agar medium and incubated at 37°C for 24 hours (20). The effect of Cu-BPDCA-Ty on gene expression was determined by a real- time PCR assay. All MRSA isolates were treated with the minimum inhibitory concentration (0.1 - 1 mg/mL) of Cu-BPDCA-Ty and incubated at 37°C for 24 hours.
3.5. RNA Extraction and cDNA Synthesis
One mL of the bacterial suspension was centrifuged and washed with DEPC-treated water. Then, microtubes were resuspended in 100 μL/mL of TE buffer (10 mmTris-HCL/1 mm EDTA, pH 8) containing 2 mg lysozyme (Sigma-Aldrich Chemie Gmbh, Steinheim, Germany). The RNA extraction was performed according to the manufacturer’s instructions (bio flux, Shibuya-ku, Tokyo, Japan). DNA contamination in the RNA preparation was digested using ‘‘RNase-free DNase I (Thermo scientific). The RNA templates were eluted in 50 μL RNase-free water. The quantity of RNA was assessed using a nanodrop spectrophotometer (Microspectrophotometer, Nano DOT, ND-3800). Purified RNA was used in a 10 μL reverse transcription reaction to synthesize cDNA. The oligo-dT and random hexamer primers were provided in prime script TM RT Reagent Kit (Takara, clontech, Japan), according to the manufacturer’s instructions.
3.6. Real Time PCR
Real- time PCR was performed in duplicate using a Real Q plus 2 x Master Mix Green (Ampliqon, Denmark) by the ABI Step one real time pcr system (Applied Biosystems). cDNA was used as a template in 20 μL reactions. The real- time PCR cycling was performed at 95°C for 15 minutes, followed by 38 cycles (95°C for 10 seconds, 60°C for 30 seconds, 72°C for 30 seconds). Finally, a melting stage was determined for the unspecific PCR product or possible primer dimmers. Water (no template) was used as a negative control in all qPCR runs. pta gene (Phosphate acetyltransferase) was used as an endogenous control. The efficiency of amplification determined standard curves based on genomic DNA by real- time PCR. The primers employed in the present study are described in Table 3. The relative expression of the mecA gene was performed using the 2-ΔΔCt method.
3.7. Statistical Analysis
Data obtained were analyzed using paired t test and one sample t test in SPSS v22 software. In all statistical calculations, P value was considered to be less than 0.05.
4. Results
4.1. Phenotypic and Genotypic Characterization of MRSA
All the 5 clinical isolates that carried nucA gene were confirmed as S. aureus. In all S. aureus isolates, except for S. aureus ATCC25923, inhibition zone for cefoxitin was < 21 mm and also they carried mecA gene, and were then identified as MRSA strains.
4.2. Synthesis of Cu-BPDCA-Ty
Synthesized Cu-BPDCA-Ty was confirmed with different techniques such as XRD and SEM analysis. The SEM (Figure 1) indicated that the compound was a uniform structure of sponge-shaped nanorods. In addition, the thicknesses of nanorods were estimated to be about less than 100 nm, but the length of the nanorods was about 500 nanometers. These data indicated the high ratio of length to width of the compound that showed this structure is appropriate for antibacterial applications (Figure 1).
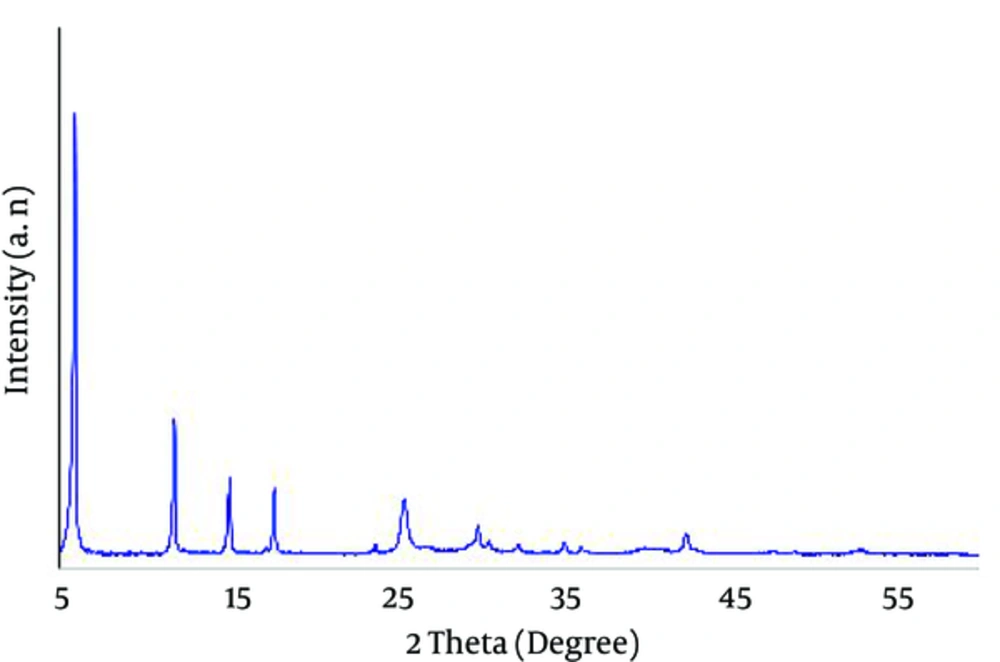
Powder XRD pattern (Figure 2), the combination of Cu-BPDCA-Ty, has indicated the lack of peaks related to copper oxide compounds (Cu2O and CuO). Generally, the detection of the crystalline structure of copper-based MOFs was disturbed by present copper oxide compounds. These data desired the successful synthesis of Cu-BPDCA-Ty. Peaks related to these intruder agents were 35.5, 38.5, 48.9, and 53.3 of CuO, and 36 and 42.5 of Cu2O. In total, MOFs are well represented as organic metal polymers or quaternary anionic polymers, with the presence of sharp and long peaks in the region of 2 Ѳ below 21 in their spectrum of XRD.
4.3. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
Minimum inhibitory concentrations and MBCs values were determined for each isolated MRSA. Results showed that the minimal inhibitory concentration required Cu-BPDCA-Ty to inhibit growth standard strain of MRSA, which was 0.5 mg/mL, and the clinical strain of MRSA, which was 0.46 ± 0.08 mg/mL. The minimal bactericidal concentrations were determined to be 0.8 mg/mL for the standard strain of MRSA and 0.7 ± 0.1 mg/mL for clinical strains of MRSA (P > 0.05).
4.4. Real- Time PCR
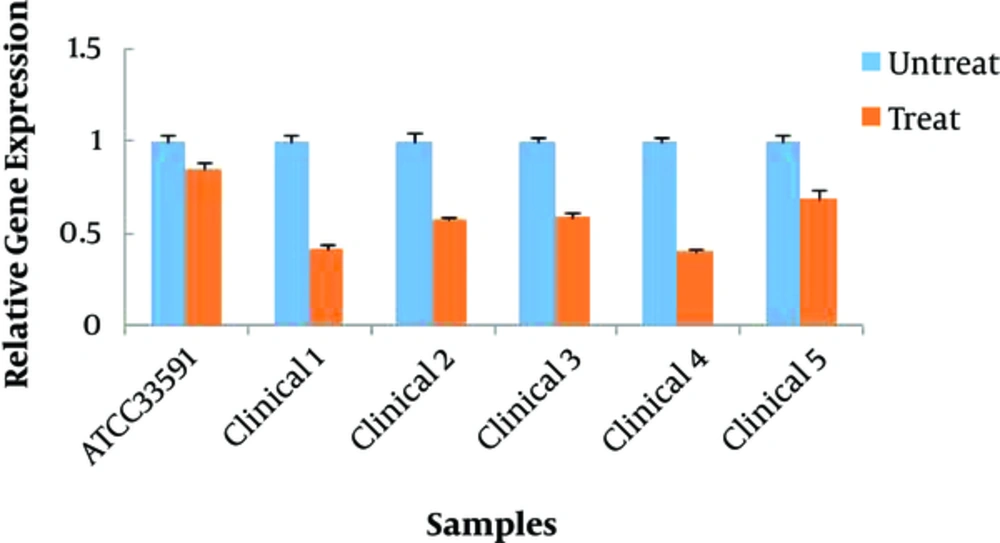
Molecular results indicated that all treated groups with Cu-BPDCA- Ty had a significant decrease in the expression of the mecA gene compared to the related control group (P < 0.05) (Figure 3).
5. Discussion
Methicillin-resistant S. aureus is an important pathogen in hospitals and communities. The emergence of multi-drug resistance of MRSA has challenged the treatment of S. aureus infection. The rapid diagnosis of MRSA is an important step in preventing the spread of infection and reducing mortality. Molecular methods are the most accurate diagnostic methods. Detection of mecA gene is a gold standard test for MRSA, which is due to the simplicity and repeatability of the molecular methods such as PCR (23). In the present study, like other similar studies, MRSA was detected using the molecular method of PCR of mecA gene, which confirms the results of previous researchers (24).
The present study revealed that MIC was different in isolated bacteria used. Our results indicated that the level of MIC was higher than other studies such as Azad et al. due to the different compounds used to build the structure of the metal organic frameworks or the greater resistance of the strains in this study (25). Metal/metal oxide nanoparticulate system, such as MOFs, have shown significant antibacterial activity. MOFs as a reservoir for metal ions lead to their gradual release and result in induced antibacterial activity (13). The antibacterial mechanism of this compound is related to the release of Cu2+ ion. These positive ions interact with the negative charge of bacterial lipids, and after destroying the wall and membrane and entering the cell, an enzymatic activity is targeted in the bacterium, which ultimately leads to a reduction in gene expression (26, 27).
Raffi et al. and Gunawan et al. attributed antibacterial activity of copper NPs to the released Cu2+ ions in bacteria and induced intracellular reactive oxygen species generation. Eventually, cytolysis of bacteria occurs because of their opposite electrical charges that lead to the release of cellular materials (28, 29). Similar to previous studies (30-33), the present study found that the antibacterial activity of nanostructures includes metal/metal oxide copper. The real- time PCR method is sensitive and commonly used to detect gene expression. The housekeeping genes (HKGs) were used for relative measurements on the change in gene expression relative to real- time PCR investigations. Ideally, the HKGs are as controls. HKG, which was utilized in the present study, was phosphate acetyl transferase (pta) gene. The expressed housekeeping gene is usually quantified at the same time. The pta gene showed good stability in the experimental settings in different conditions, such as temperature, pH, and aeration (21).
The results of the present study showed that pta gene is suitable for normalization of the mec A gene expression under the effect of the Cu-BPDCA- Ty. A significant difference existed in the expression of mecA gene between standard and clinical strains after treatment with Cu-BPDCA- Ty at concentrations of MIC. Previous studies have shown similar results, in which inhibited the expression of the resistant mecA gene. Lee et al. explained that hexane and chloroform fractions of Salvia miltiorrhiza Bunge lead to inhibition of expression of resistant genes mecA, mecI, and mecR1 (34). Chovanova et al. stated that the combination of the essential oil from S. sclarea influenced the inhibition expression of the mecA gene (35).
The essential oil similar to copper affects cytoplasmic membrane. You et al. postulated that Rhus javanica extract inhibited the genetic expression of virulence factors, such as mecA, sea, agrA, and sarA in MRSA, which may depend on the presence of phenolics in the extract (22). Similar results, indicated that the inhibition of genes expression of mecA, sea, agrA, and sarA in MRSA by Artemisia princeps, described by Choi et al. may depend on organic acids and glycosides as the major components in the A. princeps extract (36). Qiu et al. stated that TiO2 nanoparticle inhibits the growth of Shewanella oneidensis and shows a significant decrease in the expression level of the pspB gene. They declared that gene expression is a tool appropriate for detecting nanoparticles toxicity mechanisms (37). Saghalli et al. stated that ZnO nanoparticle reduced the rate of hemolysis and the hla gene expression of S. aureus. The researchers suggested that further studies use these nanoparticles as antibacterial coating for the artificial instrument to control bacterial infections (38).
6. Conclusion
In summary, the results of the present study illustrated the potential of Cu-BPDCA-Ty as a combination antibacterial against MRSA and also induced the downregulation of expression of mecA gene, which ultimately leads to the decrease of resistance to beta-lactams antibiotics. Therefore, further studies should be conducted to identify action mechanisms and toxicity of active compounds. Nanomaterial is expected to be recognized as a natural source for the development of new antibacterial components.