1. Background
Access to hygienic foods is an important factor in ensuring health. Unconfident food containing pathogenic agents cause more than 300 diseases worldwide (1, 2). Foodborne diseases root around 80 million sicknesses, 330,000 hospitalizations, and 6000 deaths in the United States annually (3, 4). Escherichia coli is a member of the huge family of Enterobacteriaceae and mostly causes food poisoning. Shiga (vero) toxin (Stx)-producing E. coli (STEC) is a sector of a substantial virulent group of E. coli entitled, enterohemorrhagic E. coli (EHEC) (4-6). Shiga toxin producing E. coli bacteria are causative agents for concentrated diseases, such as diarrhea (bloody and non-bloody), hemolytic uremic syndrome (HUS), hemorrhagic colitis (HC), and thrombotic thrombocytopenic purpura (TTP) (4-6).
Shiga toxin producing E. coli strains harbored several types of virulence factors and especially Shiga toxins (stx1 and stx2), intimin (eaeA), and hemolysin (hlyA). These genes are causative factors of settlement, adhesion, and attack of STEC bacteria to gastrointestinal mucosa (4-6). Shiga toxin producing E. coli bacteria represents an extensive prevalence of resistance to numerous clusters of antibiotics, especially quinolone, fluoroquinolone, cephalosporin, macrolide, aminoglycoside, sulfonamide, and tetracycline (4-12).
2. Objectives
Considering the indeterminate impact of STEC bacteria in milk, meat, and vegetable samples, this study aimed at assessing the frequency of virulence genes and phenotypic characterization of antibiotic- resistant STEC bacteria recovered from raw meat, milk, and vegetable samples in Iran.
3. Methods
3.1. Ethical Considerations
This study was approved by the ethical commission of enquiry adjutancy of the Islamic Azad University, Karaj Branch, Karaj, Iran (Agreement Ref Number 95-20208). This enquiry and the certificates associated with sampling were approved by Professor Afshin Akhondzadeh Basti and Dr. Valiaollah Koohdar (Endorsement Ref Number Vet-95-33711).
3.2. Samples Collection
From March to May 2015, a total of 280 samples including raw meat (n = 100), raw milk (n = 100), and vegetable (n = 80) samples were collected from the supermarkets and butcheries of Tehran province, Iran. All samples were directly moved to the laboratory using ice packs. All food samples showed normal physical properties.
3.3. Escherichia coli Isolation
A total of 10 grams of samples were standardized for 2 minutes in 90 mL of peptone water (Merck, Germany). Escherichia coli isolation and identification were done using the method described previously by Dehkordi et al. (2014) (6).
3.4. PCR Approval of Escherichia coli Strains
Escherichia coli isolates were approved again using polymerase chain reaction (PCR), according to the technique labelled by Woo et al. (2001) (13).
3.5. PCR Amplification of STEC Virulence Genes
Table 1 demonstrates the sequence of primers and PCR programs applied for identifying virulence factors (6, 14, 15). Eppendorf Flexrcycler2 (Eppendorf, Germany) was also applied. Electrophoresis was done using agarose gel (1.5%) and 1X TBE buffer strained using SYBR Green (Fermentas, Germany). Electrophoresis was done at 80 - 90 V for at least 30 minutes. Positive DNA samples taken from our previous works were applied as positive controls.
| Target Gene | Primer Sequence (5' - 3') | PCR Product, bp | PCR Programs | PCR Volume, 50 µL |
|---|---|---|---|---|
| stx1 | F: AAATCGCCATTCGTTGACTACTTCT | 366 | 1 cycle: 95°C - 3 min | 5 µL PCR buffer 10X; 2 mM Mgcl2; 150 µM dNTP (Fermentas); 0.75 µM of each primers F and R; 1.5 U Taq DNA polymerase (Fermentas); 3 µL DNA template |
| R: TGCCATTCTGGCAACTCGCGATGCA | 34 cycle: 94°C - 60 s | |||
| stx2 | F: CGATCGTCACTCACTGGTTTCATCA | 282 | 56 °C - 45 s | |
| R: GGATATTCTCCCCACTCTGACACC | ||||
| eaeA | F: TGCGGCACAACAGGCGGCGA | 629 | 72 °C -60 s | |
| R: CGGTCGCCGCACCAGGATTC | ||||
| ehly | F: CAATGCAGATGCAGATACCG | 432 | 1 cycle: 72°C -10 min | |
| R: CAGAGATGTCGTTGCAGCAG |
3.6. Antimicrobial Analysis
Susceptibility of STEC bacteria was assessed according to the principles of Clinical and Laboratory Standards Institute guidelines (16). Tetracycline (30 u/disk), ampicillin (10 u/disk), cefotaxime (30 µg/disk), gentamycin (10 µg/disk), ciprofloxacin (5 µg/disk), amikacin (30 u/disk), imipenem (30 u/disk), cotrimoxazole (30 µg/disk), enrofloxacin (5 µg/disk), sulfamethoxazole (25 µg/disk), trimethoprim (5 µg/disk), streptomycin (10 µg/disk), and chloramphenicol (30 µg/disk) antibiotic agents (Oxoid, UK) were used for this purpose.
3.7. Statistical Analysis
Chi-square test and SPSS/16.0 software were used for statistical analysis. Statistical significance was set at < 0.05.
4. Results
Table 2 characterizes the prevalence of E. coli in numerous kinds of samples. In this study, 59 out of 280 (21.07%) food samples were contaminated with E. coli strains. Vegetables (31.25%) had the highest prevalence of E. coli, while raw meat (14%) had the lowest. Statistically significant differences were observed in the frequency of E. coli in numerous samples (P < 0.05). Table 3 demonstrates the frequency of virulence genes in E. coli subtypes. Frequency of AEEC and EHEC cluster in the raw meat, raw milk, and vegetable samples was 25% and 37.50%, 16.66% and 33.33%, and 33.33% and 22.22%, respectively. Positive EHEC bacteria for stx1, eae, and ehly genes were determined as O157 serogroup.
| Types of Samples | No. Samples Collected | Positive Strains | PCR Confirmation |
|---|---|---|---|
| Raw meat | 100 | 14 (14) | 14 (14) |
| Raw milk | 100 | 20 (20) | 20 (20) |
| Vegetable | 80 | 25 (31.25) | 25 (31.25) |
| Total | 280 | 59 (21.07) | 59 (21.07) |
aValues are expressed as No. (%).
| Samples (No. Positive) | Subtypes | No. Positive Samples | Virulence Genes |
|---|---|---|---|
| Raw meat (14) | Non detected | 3 (37.50) | - |
| EHEC | 2 (25) | stx1, eae, ehly: 2 (100) | |
| AEEC | 3 (37.50) | stx1: 3 (100) | |
| stx2: 1 (33.33) | |||
| eaeA: 2 (66.66) | |||
| stx1, eaeA: 1 (33.33) | |||
| stx2, eaeA: 1 (33.33) | |||
| stx1, stx2, eaeA: 1 (33.33) | |||
| Total | 8 (57.14) | - | |
| Raw milk (20) | Non detected | 6 (50) | - |
| EHEC | 2 (16.66) | stx1, eae, ehly:2 (100) | |
| AEEC | 4 (33.33) | stx1: 4 (100) | |
| stx2: 2 (50) | |||
| eaeA: 3 (75) | |||
| stx1, eaeA: 2 (50) | |||
| stx2, eaeA: 1 (25) | |||
| stx1, stx2, eaeA: 1 (25) | |||
| Total | 12 (60) | - | |
| Vegetable (25) | Non detected | 8 (44.44) | - |
| EHEC | 6 (33.33) | stx2: 2 (50) | |
| eaeA: 3 (75) | |||
| stx1, eaeA: 2 (50) | |||
| stx2, eaeA: 1 (25) | |||
| stx1, stx2, eaeA: 1 (25) stx1, eae, | |||
| ehly:6 (100) | |||
| AEEC | 4 (22.22) | stx1: 4 (100) | |
| Total | 18 (72) | - |
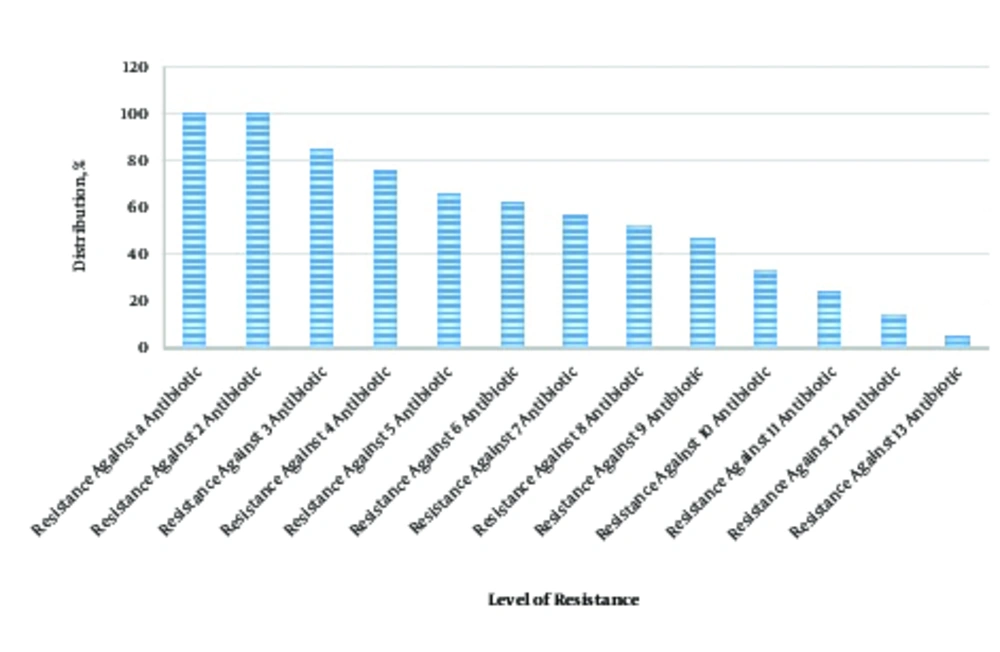
Table 4 presents the antibiotic resistance pattern of the STEC bacteria recovered from numerous types of samples. STEC bacteria revealed the vastest prevalence of resistance against ampicillin (100%), gentamycin (90.47%), tetracycline (85.71%), and ciprofloxacin (71.42%). Statistically substantial differences were observed in the incidence of resistance among different types of samples (P < 0.05). Figure 1 demonstrates the occurrence of multidrug resistance in the STEC bacteria. Shiga toxin producing E. coli bacteria were resistant to at least 2 antibiotics (100%), however, the incidence of resistance against 10, 11, 12, and 13 antibiotics was 33.33%, 23.80%, 14.28%, and 4.76%, respectively.
| Types of Samples (No. STEC Strains) | Antibiotic Resistance Pattern | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tet30 | Am10 | Cef30 | Gen10 | Cip5 | Amk30 | Imp30 | Cot30 | En5 | Sul25 | Tri5 | S10 | C30 | |
| Raw meat (5) | 4 (80) | 5 (100) | 2 (40) | 5 (100) | 4 (80) | 3 (60) | - | 2 (40) | 4 (80) | 3 (60) | 3 (60) | 2 (40) | - |
| Raw milk (6) | 5 (83.33) | 6 (100) | 3 (50) | 5 (83.33) | 5 (83.33) | 4 (66.66) | 1 (16.66) | 3 (50) | 3 (50) | 4 (66.66) | 4 (66.66) | 3 (50) | - |
| Vegetable (10) | 9 (90) | 10 (100) | 4 (40) | 9 (90) | 6 (60) | 4 (40) | 1 (10) | 3 (30) | 6 (60) | 7 (70) | 5 (50) | 4 (40) | 2 (20) |
| Total (21) | 18 (85.71) | 21 (100) | 9 (42.85) | 19 (90.47) | 15 (71.42) | 11 (52.38) | 2 (9.52) | 8 (38.09) | 13 (61.90) | 14 (66.66) | 12 (57.14) | 9 (42.85) | 2 (9.52) |
Abbreviations: amk30, amikacin (30 u/disk); am10, ampicillin (10 u/disk); cef30, cefotaxime (30 µg/disk); c30, chloramphenicol (30 µg/disk); cip5, ciprofloxacin (5 µg/disk); cot30, cotrimoxazole (30 µg/disk); en5, enrofloxacin (5 µg/disk); gen10, gentamycin (10 µg/disk); imp30, imipenem (30 u/disk); s10, streptomycin (10 µg/disk); sul25, sulfamethoxazole (25 µg/disk); tet30, tetracycline (30 u/disk); tri5, trimethoprim (5 µg/disk).
aValues are expressed as No. (%).
5. Discussion
Results of the present study revealed that virulent STEC strains of meat, milk, and vegetable samples had a considerable prevalence rate (21.07%) and high levels of antibiotic resistance against more than one antibiotic used mainly for treatment and control of food poisoning. A conceivable description for the higher frequency of STEC strains in vegetables is due to the close contact of vegetables with polluted soil, water, and animal and/or human-based mature. Another reason for this finding may be the structure of vegetables, which can facilitate the survival of bacteria. Therefore, bacteria can easily grow and increase in vegetables away from any disturbing factors such as sunlight, acidic PH, high and low temperature, low moisture, activated water, and even presence of competitor flora. Previous investigations showed that vegetables, especially lettuce, are the main reservoir of STEC strains (17-20).
The most important reasons for the higher prevalence of STEC strains in milk than meat is the fact that raw milk is mainly taken from the udder of the cows by workers’ hands. Maintaining milk in cool temperature (below 4°C) for proper safety. STEC strains are one of the most important causes of mastitis in dairy cattle. Many cases of mastitis are subclinical, and workers of milking halls are able to identify the subclinical mastitic milk. Therefore, subclinical mastitic milk samples will be collected instead of healthy milk. Momtaz et al. (2012) found that out of 268 milk samples, 73 (27.23%) were positive for E. coli, which was similar to our finding. They showed that raw milk had a high potential for survival of STEC strains (5, 21). We also found that the type of sample is a determining factor in statistical analysis of data collected from each part of the study. On the other hand, we observed a statistically significant difference in the frequency of E. coli and numerous samples (P < 0.05) and also in the incidence of antibiotic resistance among different types of samples (P < 0.05). Additionally, differences in the prevalence of E. coli were significant among milk, meat, and vegetable samples. Similarly, there were differences in the prevalence of antibiotic resistance among the E. coli strains recovered from milk, meat, and vegetable samples.
We found that 14% of raw meat samples were contaminated with E. coli. Possible explanations for this finding may be the transmission of E. coli from offal discharge, blood, floor of slaughter house, other contaminated carcasses, and also hands of meat inspectors and butchers during the process of slaughter in slaughterhouses and also transmission of bacteria during storage, transport, and maintenance of the meat. Momtaz et al. (2013) indicated that 29.02% of various types of raw meat samples were contaminated with E. coli, which was higher than our findings (4). They showed that meat, especially bovine, ovine, and caprine samples, are reservoir of virulent and resistant STEC strains.
In this study, we also focused on the virulence gene profile of the STEC strains isolated from meat, milk, and vegetable samples. We found that stx1 and eaeA were the most commonly detected virulence factors. In addition, the number of strains, which harbored all these genes (stx1, stx2, eaeA, and hly) together, was high. High presence of stx1, stx2, eaeA, and ehly genes in subtypes guarantees their considerable pathogenicity. Concurrent attendance of different virulence genes in E. coli strains specified a significant public health issue. Concurrent occurrence of Shiga toxins and intimin has been described before (22-26). In a survey accompanied by Momtaz et al. (2012), the incidence of AEEC and EHEC clusters was 49.31% and 15.06%, respectively (21). Higher frequency of AEEC subtypes was described by numerous surveys (22-26). STEC bacteria exhibited severe prevalence of resistance to tetracycline, ampicillin, gentamycin, and ciprofloxacin. Also, the level of resistance against multiple antibiotics was very high in STEC strains in our study. High prevalence of resistance against human-based antimicrobial agents in tested food samples in our study has indirectly confirmed the transmission of STEC strains from the meat inspectors and butchers, staffs of the milking halls, and human manure to meat, milk, and vegetable samples.
Frequency of resistance to chloramphenicol in the STEC bacteria of our investigation was 9.52%. There were no positive results in raw meat and milk samples. Chloramphenicol is a forbidden antibiotic. The high presence of resistance against chloramphenicol displayed its lopsided and illegal use in veterinary and, particularly, poultry science. Using poultry-based manure for growth and reinforcement of agricultural soils used for planting vegetables is the main factor that facilitates the transmission of chloramphenicol-resistant strains of E. coli to the vegetables. Highly irregular prescription of antibiotics in medicine and veterinary fields causes high prevalence of antibiotics resistance. Therefore, antibiotic resistance will appear in a very short period of time. Comparable conclusions have been described by Momtaz et al. (2012), Momtaz et al. (2013), Dehkordi et al. (2014), Rasheed et al. (2014), Colello et al. (2015), and Miles et al. (2006) (5, 6, 21, 27-29).
Momtaz et al. (2013) indicated that the maximum resistance level of STEC bacteria of meat samples was to penicillin and that resistance against ciprofloxacin and nitrofurantoin was low (4). Momtaz et al. (2012a) showed that the STEC bacteria of raw milk samples displayed severe resistance to penicillin (100%) and tetracycline (57.44%), while resistance to cephalothin (6.38%) was rare (5). Hemmatinezhad et al. (2015) described that the highest level of antibiotic resistance in the STEC bacteria of poultry meat samples belonged to gentamicin, tetracycline, and ampicillin (15). Kim and Woo (2014) found that E. coli strains, which were recovered from vegetables, exhibited rare resistance, except for cephalothin (71%). They showed that prohibiting the use of human and animal manure is the main factor for the low prevalence of antibiotic resistance of E. coli strains isolated from organic vegetables (30). High prevalence of foodborne pathogens and their considerable levels of antibiotic resistance had also been described previously from different parts of Iran (31-37).
Significant differences in the prevalence of STEC strains and the pattern of antibiotic resistance between our results and those of other investigations may be due to difference in types of samples, number of samples collected, method of sampling, method of experiment, place of sampling, weather and climate of geographical zone of sampling, opinion of medical and veterinary practitioners in antibiotic prescription, costs of antibiotics and their availability. Differences in the levels of health care and hygiene, detailed control of antibiotic prescription, accuracy of meat inspection at the slaughterhouse, hygiene of the milking halls, application of animal and human manure for fertilization of the agricultural soils used for planting vegetables, and finally the levels of personal hygiene were other factors which might have affected the prevalence of bacteria and antibiotic resistance. Moreover, considerable prevalence of E. coli and other pathogenic bacteria and their high antibiotic resistance have been described previously (38-49).
6. Conclusions
Virulent and multidrug- resistant STEC strains had a considerable prevalence in meat, milk, and vegetable samples of Iranian markets. Judicious prescription of antibiotics and accurate monitoring of meat inspection in slaughterhouses, milking in milking halls, and washing vegetables can reduce the risk of virulent and resistant STEC strains. Careful prescription of imipenem and cotrimoxazole, according to the results of disk diffusion, can help treat cases of STEC infections, especially in patients who have consumed contaminated meat, milk, and vegetables. Properly washing vegetables, cooking meat, and boiling milk, or using pasteurized milk can reduce the risk of STEC food poisoning.