1. Background
Staphylococcus epidermidis is the most important member of coagulase-negative staphylococci (CoNS) and a commensal bacteria, which is isolated prevalently from human epithelia (1). Staphylococcus epidermidis is part of the human epithelia microflora and usually has a benign relationship with the host. Colonization with S. epidermidis contributes to the maintenance of a healthy skin flora by competition with potentially harmful microorganisms (2). In the recent decades, however, with increasing use of indwelling or implanted medical devices and the increase of multi-morbid, elderly, and immunocompromised patients, S. epidermidis has emerged as an important opportunistic pathogen responsible for hospital-acquired infections, especially biofilm-associated infections (1).
Staphylococcus epidermidis is one of the often encountered biofilm-producing bacteria and the establishment of S. epidermidis as a nosocomial pathogen mainly depends on biofilm formation (2). Polysaccharide intercellular adhesin (PIA) encoded by icaADBC operon is a mediator of biofilm formation in S. epidermidis (3). Arginine catabolic mobile element (ACME), a novel genomic island, can help this species colonize the human skin, mucosal surfaces, and in-dwelling medical devices (4). Furthermore, increasing resistance rates to clinically available antimicrobial agents are an even greater problem for S. epidermidis, which limits the therapeutic options (1, 5). Mupirocin is an important antibiotic for skin and soft tissue infections (SSTIs) and the eradication of staphylococci colonization by binding to the bacterial isoleucyl-tRNA synthetase enzyme and inhibiting protein synthesis (6). However, with the increasing use of mupirocin, low- and high-level mupirocin resistance among staphylococci isolates has been increasing (7).
Fusidic acid (FA) is a valuable alternative to vancomycin for infections caused by multi-drug resistant staphylococci, especially MRSA infections (8-10). However, there is a significant trend towards increased FA resistance among staphylococci with increased duration of use. Molecular typing of S. epidermidis isolates associated with nosocomial infections has shown considerable clonal diversity and is much less studied than that for S. aureus (11). Multi-locus sequence typing (MLST), based on nucleotide sequencing of seven housekeeping genes, revealed distinct related clones of Methicillin-Resistant S. epidermidis (MRSE) clinically significant isolates and showed a worldwide predominance of only a few hospital-associated epidemic clonal lineages (11). Clonal complex 2 (CC2) was a major genetic lineage among the population structure of hospital-acquired S. epidermidis, worldwide (12, 13). Limited information is available on the resistance of S. epidermidis clinical isolates to mupirocin and FA. The aim of the present study was to investigate the resistance rate of S. epidermidis isolates from hospitalized patients to mupirocin and FA.
2. Objectives
The present study aimed at determining mupirocin and FA resistance for controlling S. epidermidis infections and eradicating staphylococci colonization.
3. Methods
3.1. Ethics Statement
The ethics committee of the first Affiliated Hospital of Wenzhou Medical University exempted this study from review, because the present study focused on bacteria (ID: 62697).
3.2. Bacterial Isolates
Seven hundred and seventy-one non-duplicate S. epidermidis isolates were collected consecutively from various specimens of inpatients from January 2012 to December 2015 at the first Affiliated Hospital of Wenzhou Medical University in Wenzhou, east China. The isolates were identified as S. epidermidis using Gram staining, catalase test, coagulase test, and VITEK automatic microbiology analyzer (bioMérieux, Marcy l’Etoile, France).
3.3. MRSE Identification
Polymerase chain reaction was used to detect whether the tested strains harbored mecA, with MRSA N315, as the positive control strain. The strains carrying mecA were defined as MRSE. Moreover, all the clinical strains were targeted for mupA by PCR assays.
3.4. Screening for Mupirocin and FA Resistance
Mupirocin and FA minimum inhibitory concentration (MIC) values for S. epidermidis isolates were determined by an agar dilution method, in accordance with the CLSI guidelines. The S. aureus ATCC29213 was used as the control strain. Staphylococcus aureus isolates with MICs of 8 to 256 mg/mL and ≥ 512 mg/mL were defined as having low- and high-level resistance to mupirocin (7). The researchers defined S. epidermidis with MIC of > 256 mg/mL as having high-level resistance to mupirocin. The interpretive criterion of FA susceptibility for staphylococci is in accordance with the European Committee for Antimicrobial Susceptibility Testing (EUCAST) / British Society of Antimicrobial Chemotherapy (BSAC) criteria (susceptible, MIC < 2 μg/mL; resistant, MIC ≥ 2 μg/mL).
3.5. Antimicrobial Susceptibility Testing for Mupirocin-Resistant Staphylococcus epidermidis Isolates
The susceptibility of the mupirocin- and/or FA-resistant S. epidermidis clinical isolates to commonly used antimicrobial agents and screening of MRSE was done using VITEK-2 compact automated microbiology analyzer platform (bioMérieux, Marcy l’Etoile, France), according to the manufacturer’s instructions. Results interpretation was in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (14). Antimicrobial agents included ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), rifampin (10 μg), tetracycline (30 μg), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). All antimicrobial disks were obtained from Oxoid Ltd., and S. aureus ATCC25923 was used as a quality control strain for antimicrobial susceptibility testing.
3.6. Detection of Mupirocin and FA Resistance Determinants
mupA, conferring high-level mupirocin resistance, was detected by PCR, as described previously (15). fusA mutations and acquired FA resistance determinants, including fusB, fusC and fusD, were detected by PCR assays with primers and reaction conditions described previously (16) and DNA sequencing.
3.7. MLST Typing
Multi-locus sequence typing for S. epidermidis isolates was performed by amplification of internal fragments of the seven housekeeping genes, including arcC, aroE, gtr, mutS, pyrR, tpiA, and yqiL, described previously (17). The PCR products of seven housekeeping genes, tested for MLST typing, were purified and sequenced. The numbers of alleles and sequence types were assigned using an online database (http://sepidermidis.mlst.net/).
3.8. Statistical Analysis
Prevalence of MRSE was analyzed using GraphPad Prism 7.0 software. Results were considered statistically significant if P-values were < 0.05.
4. Results
4.1. Prevalence of Mupirocin Resistance Among S. epidermidis Clinical Isolates
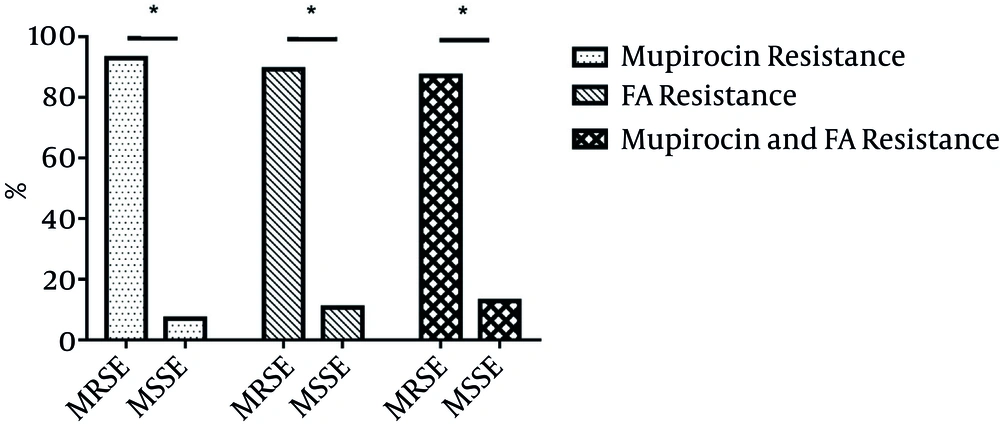
Among 711 S. epidermidis clinical isolates, 71 (9.99%) with mupirocin MICs ranging from 16 to > 256 mg/L were found to be resistant to mupirocin. Low- and high-level mupirocin resistance proportions were 0.84% (6/711) and 9.14% (65/711), including 71 mupirocin-resistant isolates, 66 (92.96%) and five (7.04%) MRSE and methicillin-susceptible S. epidermidis (MSSE) (Figure 1). In the present study, more than 50% of resistance for 71 mupirocin-resistant isolates to non-β lactam included erythromycin (84.51%, 60/71), clindamycin (74.65%, h53/71), ciprofloxacin (63.38%, 45/71), gentamicin (67.61%, 48/71) and trimethoprim-sulfamethoxazole (71.83%, 51/71), while the resistance rates to rifampin (39.44%, 28/71) and tetracycline (18.31%, 13/71) were less than 50% (Table 1).
| Antibiotics | Mupirocin-Resistant Isolates (N = 71) | FA-Resistant Isolates (N = 56) |
|---|---|---|
| Mupirocin | 71 (100) | 31 (55.36) |
| FA | 31 (43.66) | 56 (100) |
| Erythromycin | 60 (84.51) | 45 (89.36) |
| Clindamycin | 53 (74.65) | 36 (64.29) |
| Ciprofloxacin | 45 (63.38) | 33 (58.93) |
| Gentamicin | 48 (67.61) | 36 (64.29) |
| Trimethoprim-sulfamethoxazole | 51 (71.83) | 37 (66.07) |
| Rifampin | 28 (39.44) | 26 (46.43) |
| Tetracycline | 13 (18.31) | 13 (23.21) |
a Values are presented as No. (%).
4.2. Prevalence of FA Resistance Among S. epidermidis Clinical Isolates
Among 711 S. epidermidis isolates, 56 (7.87%) with FA MICs ranging from 4 to 32 mg/L were resistant to FA. Fifty (89.29%, 50/56) and six (10.71%, 6/56) FA-resistant isolates were MRSE and MSSE (Figure 1). In the present study, FA-resistant isolates exhibited more than 50% of resistance rates to erythromycin (89.36%, 45/56), clindamycin (64.29%, 36/56), ciprofloxacin (58.93%, 33/56), gentamicin (64.29%, 36/56), and trimethoprim-sulfamethoxazole (66.07%, 37/56), yet less than 50% of the resistance was towards rifampin (46.43%, 26/56) and tetracycline (23.21%, 13/56) (Table 1).
4.3. Prevalence of Both Mupirocin and FA Resistance Among S. epidermidis Clinical Isolates
Of 711 S. epidermidis clinical isolates, 31 (4.4%) were resistant to both mupirocin (MICs > 256 mg/L) and FA (MICs ranging from 4 - 16 mg/L). The prevalence of both mupirocin and FA resistance among mupirocin- and FA-resistant isolates was 43.66% (31/71) and 55.36% (31/56). Of 31 isolates with both mupirocin and FA resistance, 27 and four were MRSE and MSSE (Figure 1). Ten and seven were isolated from blood and catheter. Thirty-one isolates were isolated from 17 wards, with four from the intensive care unit (ICU). Resistance to both mupirocin and FA was found among S. epidermidis isolates.
4.4. Detection of Mupirocin and FA Resistance Determinants
In the present study, as shown in Table 2, all six S. epidermidis isolates with low-level mupirocin resistance were negative for mupA. However, 23 (35.38%) of 65 isolates with high-level resistance to mupirocin were found to carry this gene. In the present study, 12 FA-resistant S. epidermidis clinical isolates were positive for fusB, while only one was positive for fusC. The fusA mutations and fusD were not found in any of the tested isolates.
| Stains | STs | Specimen | FA (mg/L) | Mupirocin (mg/L) | mupA | fusB | Ward | Year |
|---|---|---|---|---|---|---|---|---|
| BP201 | Drainage | 4 | > 256 | - | - | Pediatric | 2012 | |
| BP202 | 23 | Drainage | 8 | > 256 | - | - | Pediatric | 2012 |
| BP204 | 2 | Sputum | 32 | > 256 | - | - | Gastroenterology | 2012 |
| BP205 | 2 | Sputum | 16 | > 256 | - | - | Respiratory | 2012 |
| BP206 | Sputum | 16 | > 256 | - | - | Neurology | 2012 | |
| BP207 | Wound exudate | 32 | > 256 | - | - | Pediatric | 2012 | |
| BP208 | 2 | Catheter | 16 | > 256 | - | - | Hepatobiliary Surgery | 2013 |
| BP209 | 125 | Cather | 16 | > 256 | - | - | Nephrology | 2014 |
| BP210 | Drainage | 8 | > 256 | - | - | Nephrology | 2014 | |
| BP211 | 2 | Urine | 16 | > 256 | - | - | Infectious disease | 2015 |
| BP212 | 2 | Blood | 16 | > 256 | - | - | ICU | 2014 |
| BP213 | 2 | Blood | 16 | > 256 | - | - | ICU | 2014 |
| BP214 | 2 | Catheter | 16 | > 256 | - | - | Dialysis | 2014 |
| BP216 | Wound exudate | 8 | > 256 | - | - | Dialysis | 2014 | |
| BP217 | 466 | Drainage | 16 | > 256 | - | - | Cardiovascular | 2015 |
| BP218 | 2 | Ascties | 16 | > 256 | - | - | Gastroenterology | 2015 |
| BP219 | 2 | Blood | 16 | > 256 | - | - | ICU | 2015 |
| BP220 | 2 | Catheter | II | > 256 | - | - | ICU | 2015 |
| BP221 | 23 | Dialysate | 16 | > 256 | - | - | Nephrology | 2015 |
| BP222 | Blood | 16 | > 256 | - | - | Dialysis | 2015 | |
| BP223 | 2 | Blood | 16 | > 256 | + | - | Comprehensive ward | 2015 |
| BP224 | 2 | Blood | 16 | > 256 | - | - | Comprehensive ward | 2015 |
| BP225 | 2 | Blood | 16 | > 256 | + | + | Comprehensive ward | 2015 |
| BP226 | Catheter | 16 | > 256 | - | - | Cardiovascular ICU | 2015 | |
| BP227 | 130 | Ascites | 16 | > 256 | - | - | Nephrology | 2015 |
| BP228 | 2 | Catheter | 16 | > 256 | + | + | Infectious disease | 2015 |
| BP229 | Blood | 16 | > 256 | - | - | Nephrology | 2015 | |
| BP230 | Blood | 16 | > 256 | - | - | Comprehensive ward | 2015 | |
| BP231 | Tissue | 16 | > 256 | - | - | Hepatobiliary Surgery | 2015 | |
| BP232 | 466 | Catheter | 16 | > 256 | - | - | Proctology | 2015 |
| BP233 | 2 | Blood | 16 | > 256 | + | - | Neurology | 2015 |
Among 31 isolates with both mupirocin and FA resistance, only two were positive for fusB, conferring FA resistance and only six were positive for mupA, conferring high-level mupirocin resistance.
4.5. Molecular Characteristics of S. epidermidis Clinical Isolates with both Mupirocin and FA Resistance
Among 31 S. epidermidis isolates with both mupirocin and FA resistance, 15, three and two belonged to ST2, ST466, and ST23, respectively (Table 2). ST125 and ST130 were found only in one isolate each. Nine isolates with different loci patterns did not match the available STs in an online database (http://sepidermidis.mlst.net/). All 15 ST2 S. epidermidis isolates were MRSE and had high-level resistance to mupirocin (MICs > 256 mg/L). The MICs of FA for 14 ST2 isolates was 16 mg/L. The resistance profiles of 15 S. epidermidis ST2 isolates was similar, with resistance to penicillin, ciprofloxacin, levofloxacin, rifampin, gentamicin and trimethoprim-sulfamethoxazole, and susceptibility to vancomycin, linezolid, nitrofurantoin, quinupristin/dalfopristin, tetracycline and teicoplanin. Twelve (80.00%) of the 15 ST2 isolates were resistant to erythromycin and clindamycin. Fifteen ST2 isolates were isolated from blood (7), catheter (4), sputum (2), urine (1) and ascites (1), and disseminated among eight wards from 2012 to 2015 (Table 2). However, all four isolates (three from blood and one from catheter) with resistance to both FA and mupirocin from the ICU belonged to ST2 (Table 2).
5. Discussion
A high prevalence (61%) of mupirocin resistance was found among CoNS isolates, collected from catheter-associated bloodstream infections in very preterm neonates (18). In vivo transfer of high-level mupirocin resistance from S. epidermidis to S. aureus was associated with failure of mupirocin prophylaxis (19). Rates of low- and high-level mupirocin resistance were 9.4% and 3.3% in S. epidermidis, reported by a multi-centre surveillance study, including 26 laboratories from Austria, Germany, and Switzerland in November 2001 (20), while in the present study this rate was 0.84% and 9.14%. Mupirocin resistance may be helpful in the spread of multidrug resistance through co-selection with other resistance genes. Previous reports found that mupirocin-resistant S. aureus isolates were multi-resistant to other antimicrobial agents, such as ciprofloxacin, clindamycin, and tetracycline (21, 22). Similarly, in present study, mupirocin-resistant S. epidermidis were also multi-drug resistant to other antimicrobial agents.
McLaws et al. reported a high prevalence (46%, 23/50) of resistance to FA in S. epidermidis clinical isolates (23). Twenty-five percent of methicillin-resistant CoNS and 15% of methicillin susceptible CoNS strains isolated from blood cultures of septicemic patients in Turkey were resistant to FA (24). Compared with reports mentioned above, the prevalence of FA in the present study was relatively low, as for the prevalence of both mupirocin and FA resistance among S. aureus clinical isolates. Doudoulakakis et al. reported that 95.2% (417/438) of mupirocin-resistant S. aureus isolates were associated with community-associated infections among children in Greece were resistance to FA (25). Park et al. reported that all 13 low-level mupirocin-resistant S. aureus isolates and five (55.6%) of nine high-level mupirocin-resistant S. aureus isolates were resistant to FA (26). Overall, 103 (15.5%) of 664 S. aureus isolates from the UK were resistant to both FA and mupirocin (high level) (27). However, both mupirocin and FA resistance among S. epidermidis isolates was not reported previously. The present study was the first report of resistance to both mupirocin and FA, among S. epidermidis isolates. Emergence of both mupirocin and FA resistance among staphylococci limits the choice of antimicrobial agents for the treatment of multidrug-resistant staphylococci infections, especially SSTIs. Both mupirocin and FA-resistant isolates were susceptible to nitrofurantoin, quinupristin/dalfopristin, linezolid, vancomycin, and teicoplanin.
A new isoleucyl-tRNA synthetase with many similarities to eukaryotic enzymes, encoded by plasmid-borne gene mupA, conferred high-level resistance in staphylococci (28). Two major FA resistance mechanisms, the alteration of the drug target site caused by mutations in fusA, encoding elongation factor G (EF-G) or rplF encoding ribosome protein L6, and the protection of the drug target site by FusB family proteins, including FusB, FusC, and FusD, were reported in S. aureus (8). In staphylococci, high-level FA resistance is usually associated with mutations in fusA, while low-level resistance is generally caused by plasmid-mediated resistance genes, including fusB, fusC and fusD (29). Colonized staphylococci on skin may be a reservoir for FA resistance genes (30). The monitor for presence of FA resistance genes among S. epidermidis should be helpful for preventing the dissemination of fusidic acid resistance. Surprisingly, low positive rate for fusB conferring FA resistance and mupA conferring high-level mupirocin resistance, indicated that new mechanisms may be associated with both mupirocin and FA resistance. Further studies are needed to investigate theses new mechanisms.
The results of multi-locus sequence typing (MLST) revealed that the S. epidermidis ST2 clone with resistance to both FA and mupirocin had disseminated in the hospital of the current study. ST2 was the predominant clone among S. epidermidis clinical isolates worldwide (12, 13). A report from China showed that 91.7% (297/324) of S. epidermidis from the community and hospital environments belonged to clonal complex 2 (CC2) (13). Furthermore, CC2 comprised 74% of the S. epidermidis isolates from 17 national centers between 1996 and 2001 (12). The majority (62/71; 87.3%) of S. epidermidis clinical isolates from US hospitals belonged to CC2 (31). The current authors speculate that acquiring resistance to both FA and mupirocin, as well as multi-resistance to other antimicrobial agents, contributes to the spread of S. epidermidis ST2 clone. ST23 was found among linezolid-resistant S. epidermidis isolates (32, 33). In the present study, ST23 was identified in two S. epidermidis isolates with resistance to both mupirocin and FA. The present study first reported that ST23 was identified among S. epidermidis isolates, isolated from China. Although ST125 and ST130 exist in online databases (http://sepidermidis.mlst.net/), literature on S. epidermidis ST125 and ST130 isolates has not been found. The present study was the first report of ST125 and ST130 among S. epidermidis clinical isolates.
5.1. Conclusions
Taken together, the present study is the first report of resistance to both mupirocin and FA among S. epidermidis isolates. Dissemination of S. epidermidis ST2 clone with both FA and mupirocin resistance can cause trouble in controlling S. epidermidis infections.