1. Background
Skin infections with dermatophytes are common human infections called dermatophytosis (1, 2). The pathogenesis of dermatophytosis is not well defined, but a characteristic feature of dermatophytes is that they can colonize the surface of skin and release enzymes that break down host's fats and proteins (3). The special ability of dermatophytes to decompose creatine and use it to produce nitrogen leads to their colonization on the surface of the skin and penetration into the host epidermis (3).
Dermatophytes are a group of keratinophilic molds with global distribution that based on the newest proposed taxonomy consist of more than 50 species in the genera of Trichophyton, Microsporum, Epidermophyton, Nannizzia, Arthroderma, Lophophyton, and Paraphyton. Dermatophytes are members of the oldest group of microorganisms recognized as human disease agents. Results of Robert Remak and David Gruby investigations in 1841 introduced the taxonomy of these fungi (4). Microsporum audouinii, Epidermophyton floccosum, Trichophyton schoenleinii, T. tonsurans, and T. mentagrophytes had already been described between 1840 and 1875 as five important dermatophyte species before the introduction of axenic culture by Louis Pasteur (5). Trichophyton rubrum is the only pervasive modern dermatophyte absent from the above-mentioned list (6), which has been theorized to have emerged in the 20th century (7).
Culture of dermatophytes and explanation of new species have enormously accelerated after Pasteur's time. Several species were defined by combining clinical pictures and morphological characters in vitro, including 16 species associated with humans that were introduced between 1870 and 1920 through Sabouraud's (8) magisterial overview of dermatophytes that set a new standard.
Application of new methodological standards throughout the following decades led to the definition of a large number of new species and recombined names. However, generic concepts remained confused, which led to frequent name changes involving 350 names around 1950. Consequently, anamorph nomenclature was stabilized by the widespread acceptance of Epidermophyton, Microsporum, and Trichophyton as three genera covering all the dermatophyte species (9). Due to their keratinophilic nature, dermatophytes, which are classified in the three genera of Microsporum, Epidermophyton, and Trichophyton infect the skin, hair, and nails and ecologically infect humans, animals, and soil, respectively. These fungi have several physiological and antigenic relationships with each other, and they are able to create a variety of clinical signs in different anatomical sites, including dermatophytosis of the scalp, body, nail, thigh, foot, faciei, hand, and beard (10).
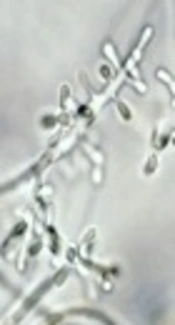
Trichophyton rubrum is the most common cause of tinea corporis in the world (11). This fungus was first discovered and explained by Malmsten in 1845 (12). Due to the fact that T. rubrum is an anthropophilic fungus, it has been less frequently found in animals (13). However, Van Breusegem believes that it is possible to create an experimental infection in Indian guinea pigs, and Rice managed to develop the disease in a rabbit that had been irradiated on the abdomen (10). The infection caused by T. rubrum can be acute and chronic, and in some people can persist for the rest of life. Thu fungus may have a period of extinction, but it can also have recurrence. In people with low anti-dermatophytic activity in serum, there is a widespread invasion to dermal and epithelial cells (14). If this fungus attacks the hair, its infection is ectothrix. Urease and hair perforation tests are negative in T. rubrum, but its absorption of sorbitol is positive. It creates red colonies on Corn Meal Agar and greenish colonies on Littman Oxgall Agar. Trichophyton rubrum lacks fluorescence and its sexual status remains unknown. In terms of morphology and colony view, it makes white slow-growing fluffy colonies with wine red reverse without changing pH on Sabouraud Dextrose Agar (SDA) with cyclohexamide, chloramphenicol, and gentamicin. Microscopically, the colorless, transparent mycelia have transverse walls. The microconidia are tear-shaped or peg-shaped and are sporadically formed around the mycelia. The macroconidia are elongated cigar-shaped, which have smooth surface with thin aligned walls and 2 - 8 septa (13).
Antimicrobial peptides (AMPs) are important molecules in natural immunity of the skin, which potentially have anti-microbial effects and quickly eliminate microorganisms (15). Since the skin is the most important organ in contact with the environment as well as the first barrier against microorganisms, AMPs are the chemical boundary between the host tissues and the environment (16-18). Human b-diphensyne (hBD-3) and RNase 7 are two important AMPs that inhibit the growth of dermatophytes in vitro (19).
EGFR is a type of mucosal tyrosine kinase that plays an important role in the differentiation, proliferation, and invasion of human cancers with an epithelial origin (20). In fact, EGFR is a regulator of deep signals and adjusts the mentioned factors (21, 22). It is reported that patients treated with suppression of EGFR will suffer from dermatophyte infections (23). Therefore, EGFR inhibition may have a negative effect on AMP localization in dermatophyte-infected keratinocytes and blockage of EGFR, resulting in a significant reduction of hBD-3 and RNase 7 in keratinocytes encountered with T. rubrum. Increasing AMP levels may also help the host in control, growth inhibition, and diffusion of other dermatophytes besides T. rubrum (24). Since there are no molecular studies on miRNAs in T. rubrum and because the expression of EGFR gene depends on factors such as the presence or absence of miRNAs, in this study, we aimed to shed light on the role of miRNAs in the expression of EGFR gene that causes the presence or absence of AMPs, resulting in the occurrence or absence of dermatophytosis with T. rubrum.
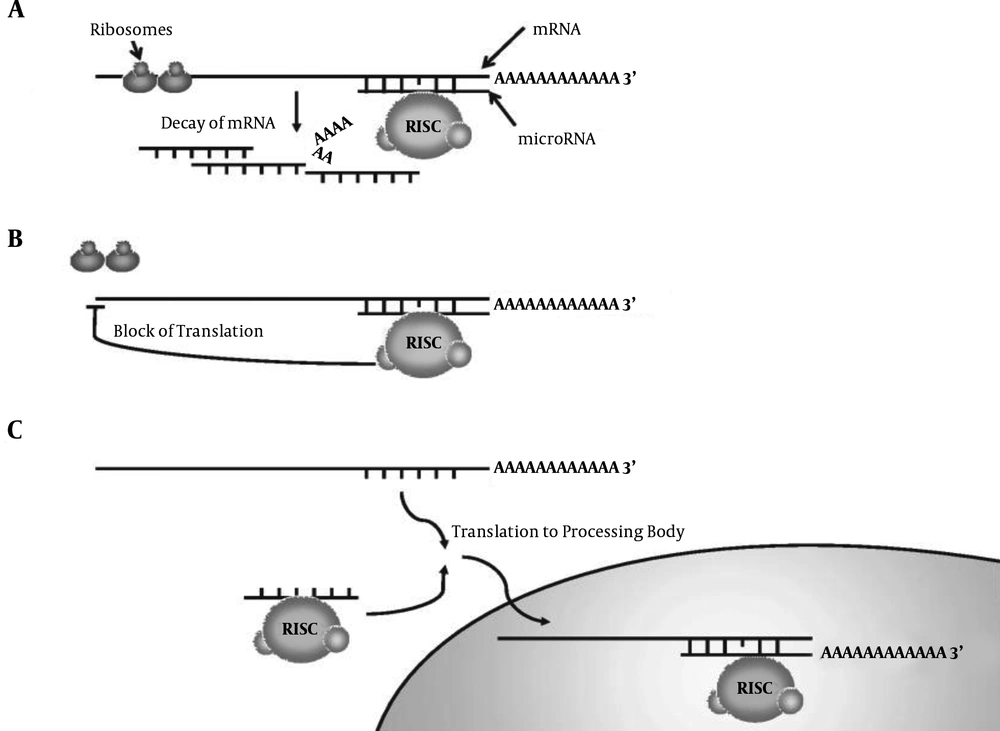
MiRNAs are non-encoding small RNAs with 18 - 26 nucleotides (25). MiRNAs are conserved molecules and intracellular regulatory RNAs that modulate gene expression via interference pathway (RNAi). RNAi is a kind of post-transcriptional shutdown mechanism in eukaryotes, which is characterized by the creation of double-stranded RNA. In this way, these molecules control gene expression after transcription by inhibiting the translation of mRNA or inducing its dissociation (26-31). There are several pathways in which miRNAs influence and inhibit the expression of their target genes by affecting mRNAs of the target genes. Two important activities of miRNA are as follows. In the first mechanism, RNA-induced silencing complex (RISC)-coupled miRNAs pair up to the autologous 3'UTR mRNA and control the gene transcription after its expression by cutting or inhibiting the target mRNA translation (Figure 1A and 1B). In another mechanism, translation inhibition occurs with mRNA enclosing in processing bodies (P-bodies; Figure 1C). Therefore, mRNA is not translated into proteins (32, 33).
Mechanism of translation inhibition of target genes by microRNA via. A, mRNA dissociation; B, inhibition of mRNA translation; C, enclosing P-bodies (34)
Bernhard's research at International Institute of Cancer in the United States on head and neck squamous cell carcinoma (HNSCC) suggested a link between increased expression of EGFR gene and miR-212 gene, since the elevation in EGFR gene expression is regulated by miR-212, which has been observed in keratinocytes and is likely to increase the risk of infection with T. rubrum (35).
2. Objectives
After extensive research by bioinformatics studies using miRWalk database on the association of EGFR gene with miRNAs, it is concluded that among the many miRNAs, miR-212 has the greatest impact on the expression of EGFR gene. Therefore, in this study, miR-212 has been nominated for consideration. Considering the approach of some of the world’s leading pharmaceutical industries to designing new miRNA-based drugs, the results of this study can be a small step towards this goal.
3. Methods
3.1. Ethics Statement
Prior to the study, we obtained the approval of the Ethics Committee of Kerman University of Medical Sciences (code: IR.KMU.REC.1394.416).
3.2. Sample Collection
The number of specimens considered in this study was 72, including 36 infected specimens with T. rubrum and 36 control specimens from healthy margins of the lesion. First, the scraping site of specimen and control was disinfected using 70% alcohol. To prepare patient samples, the skin around the lesion infected with dermatophyte (especially new parts in which the fungus was active and had satellite growth) was shaved with a scalpel and placed in a cryotube. To provide the control sample, the margin of healthy skin at a distance of 15 - 20 cm from the lesions of dermatophyte infection patients was shaved using a scalpel and put into a cryotube.
Considering that our research was on skin samples, we focused on dermatologic specimens and ignored nail and hair samples. The samples were mostly from lesions of tinea corporis, tinea cruris, and tinea pedis. Since the relationship between dermatophytosis and patient conditions was not important in this study, the physiologic and pathologic conditions of the patients were not considered. Since a -70°C freezer was not available at the sampling site, the specimens were poured into cryotubes and transferred into liquid nitrogen (-196°C).
Urease, pigmentation on Cornmeal agar, and hair perforation tests were used to differentiate between T. rubrum and T. mentagrophytes due to their high clinical similarity and colony appearance. Slide culture technique was performed to observe the fungal reproductive system. The use of molecular methods like sequencing and RFLP is advantageous to distinguish between dermatophytes such as T. rubrum and T. mentagrophytes (36); however, we used direct observation and culture of fungi for this purpose.
3.3. RNA Isolation and Real-Time PCR Testing
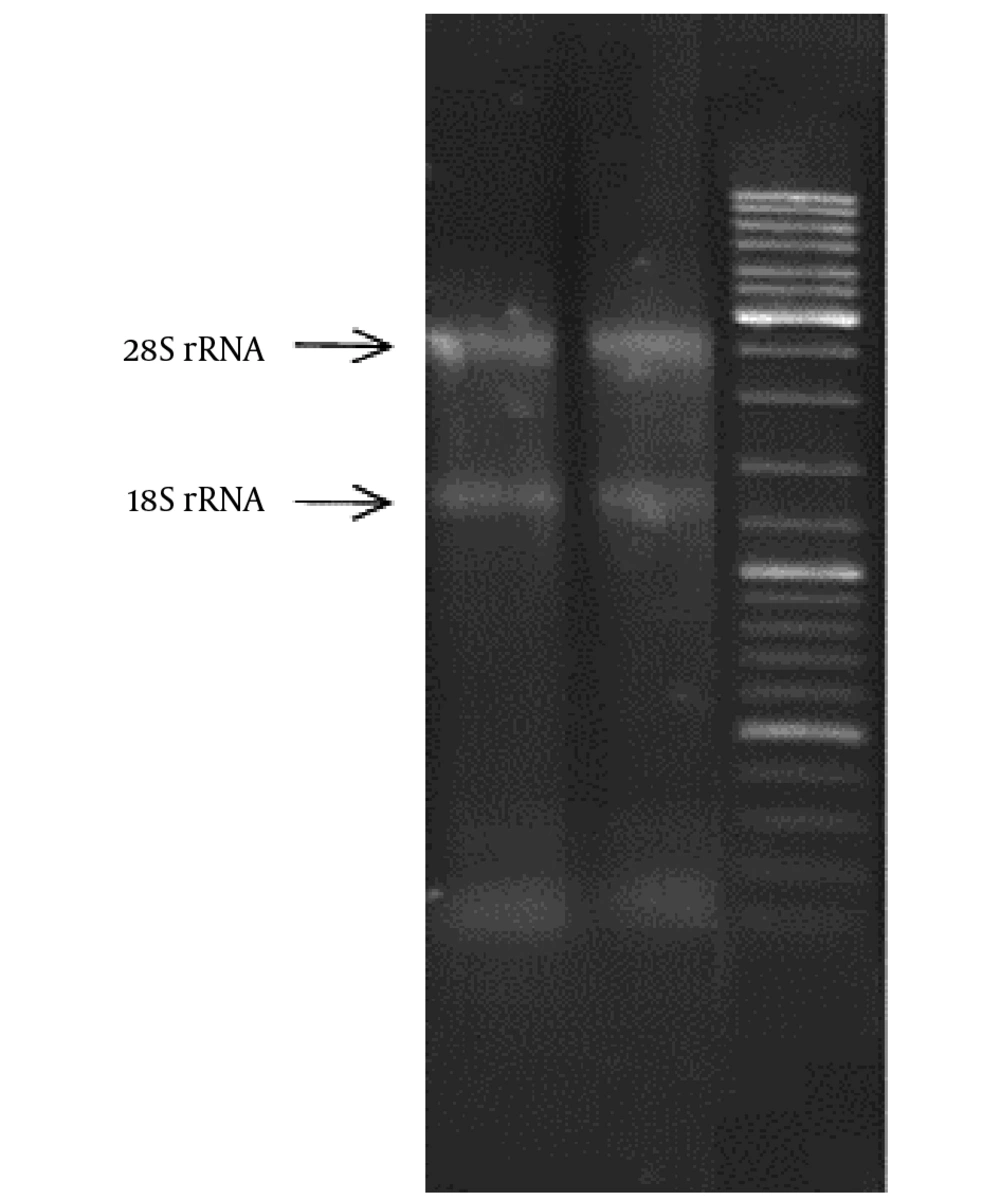
Using RNX-Plus kit (CinnaGen, Iran), RNA was isolated during 18 steps. Then, using a nano-drop device via a quantitative method, the purity degree of RNA samples was obtained through 260 - 280 nm absorbance. The results obtained from this step indicated that the isolated RNA could be used with high confidence in the next stages of the study. The use of DNase I enzyme and DNase treatment process was obligatory to remove those parts of genomic DNA that were not removed during RNA isolation. In this study, DNase I (RNase free) kit (CinnaGen, Iran) was used. In addition, 1% agarose gel was employed to evaluate the quality of the isolated RNA and maintain its integrity. An RNA sample, which is undisturbed chemically and has standard biological quality, shows a specific bonding pattern on agarose gel. The presence of S18 and S28 ribosomal RNA bands represents healthy and intact RNA. cDNA synthesis kit (Takara, Japan) was used in reverse transcription reaction for EGFR gene cDNA synthesis via an mRNA and its optimization. In reverse transcription reaction for miR-212 gene cDNA synthesis via an mRNA, cDNA synthesis kit was used (Exiqon, Denmark). Both kits were matched to 2-step real-time polymerase chain reaction (RT-PCR) System. The primers used in RT-PCR are presented in Table 1.
Primer Sequences Used in the Real-Time Polymerase Chain Reaction
The synthesis of EGFR and GAPDH gene primers was undertaken by CinnaGen Company, but miRNA primers were prepared by Exiqon Company. To carry out the 2-Step RT-PCR test (a double-test for each sample) for EGFR and GAPDH genes, we used the Ampliqon kit (Virazhen, Denmark), and according to the kit manual, the Light Cycler *96 device was used. Mercury LNA Universal RT microRNA PCR kit (Exiqon, Denmark) was applied to conduct 2-Step RT-PCR test (a double-test for each sample) for miR-212 and miR-103a-3p, and Light Cycler *96 device was used for miR-212 genes according to the kit's manual.
3.4. Data Aggregation and Analysis
Expression of genes was calculated by 2-ΔΔCt formula in Excel software. Quantitative data were described using mean and standard deviation, and qualitative or categorical data were described with frequency distribution tables. To analyze the data, correlation coefficients and, if necessary, paired test were used, and P value less than 0.05 was considered significant. RT-PCR data analysis software was also used. The statistical analyses were performed in SPSS version 21 with 95% confidence interval. The 2-ΔΔCt Formula is as follows:
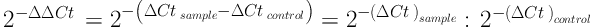
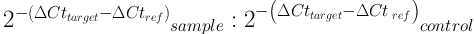
Cttarget= Mean Ct for target gene; sample = treated samples; Ctref = Mean Ct for reference gene; control = control samples; Ct (cycle threshold) indicates the cycle in which RT-PCR products surpass a threshold, which is used as a number in calculations.
4. Results
After culture of skin lesions' scales on SDA with cycloheximide and chloramphenicol, white, smooth to protuberant fluffy colonies of T. rubrum were grown with wine red reverse pigmentation. To confirm the observed fungus, we prepared a slide culture from all the samples. The slides and coverglass obtained from slide culture were stained with Lactophenol Cotton Blue. The reproductive organs of all the samples were observed using × 40 microscopy as colorless, transparent, and delicate mycelia with transverse wall, tear-shaped and cigar-shaped macrocondia with 2 - 8 septa (39). After isolating total RNA of the cells, the concentration of each sample was determined. A260/A280 ratio was also calculated for each sample, and the obtained values for the samples were read between 1:7 and 1:9 by the device, which indicated the acceptable purity of RNA. In addition, the results of isolated RNA in terms of qualitative analysis showed that S28 and S18 bands and S5 ribosomal RNA were clearly observed on 1% agarose gel for each sample (Figure 2).
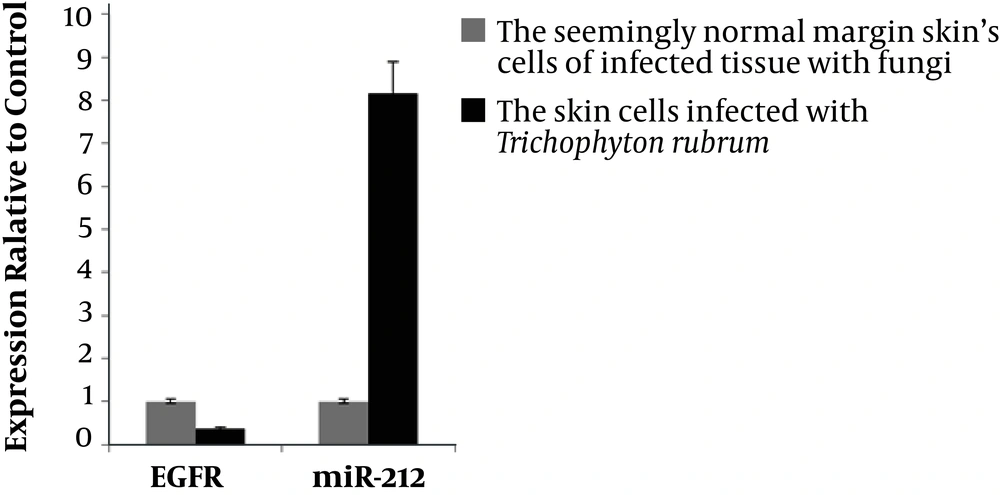
The amplification curve of internal control or reference genes used in this study, which were called GAPDH and miR-103a-3p with the discussed genes (i.e., EGFR and miR-212), are shown in the figure below. At the end of the study, comparison of the expression of target genes from dermatophytosis spots and their healthy margins indicated significant changes, P = 0.0001, using 2-ΔΔCt formula via Excel software (Figure 3).
5. Discussion
Research on effects of negative induction of miRNAs on EGFR gene, which result in gene breakdown and cause a variety of diseases particularly in the field of cancers, has been well documented, including Bernhard's extensive research at International Institute of Cancer in the United States of America on HNSCC. The mentioned study suggested that the increased expression of miR-212 gene results in suppression of EGFR gene-related mRNAs and ultimately, reduces the expression of this gene (35). It was the first research to show the important role of miRNA in regulating the ligand of the gene receptor.
Several years later, the detection of miRNAs in cancers became important, and it became clear that receptor ligands of genes were regulated by miRNAs that were reported earlier (40-42). Bernhard found that miR-212 had a specific role in regulating EGFR and identified EGFR gene levels at different levels of the cells involved with HNSCC. Reduced expression of miR-212 gene was demonstrated by increased expression of EGFR gene (43). Therefore, in their study, the association between miR-212 and EGFR genes was clearly expressed. In another study, Yasemin Helene Firat explained the presence or absence of AMPs in keratinocytes of the skin by EGFR gene expression and the way to deal with skin infections. Thus, increased expression of AMPs was the most important defense of keratinocytes against microorganisms (15, 16, 24).
Her study clarified that keratinocytes are the first cells in the face of the common dermatophytosis by T. rubrum (14, 44). They synthesized IFN-γ and IL-17A cytokines, which increased the presence of AMPs in keratinocytes (45). Finally, AMPs found in keratinocytes (including hBD-3 and RNase7) had the potential to cope with T. rubrum (19). Since the inhibition of EGFR gene negatively affects the presence of AMPs, it often leads to colonization of dermatophytes on the skin, which leads to dermatophytosis (23). According to the two mentioned studies that were relevant to our research, we also seek to determine the relationship between EGFR and miR-212 genes with regard to T. rubrum infection, as a common factor in the development of dermatophytic diseases in the world. According to the analysis, miR-212 expression was changed in skin cells after infestation with T. rubrum. In the scales of fungal lesions and healthy skin cells around them, an increase and decrease in the expression of this miRNA occurred, respectively. miRNAs control the expression of their target genes via affecting the target genes' mRNA.
It should be noted that a specific miRNA molecule does not have a specific target mRNA functionally; in other words, a miRNA may have more than one target mRNA molecule. Conversely, this also holds true in that each molecule of mRNA can affect several miRNAs (32, 33) .It should also be noted that a large number of molecular targets of miR-212 in different cells, such as skin cells, have not yet been verified in vitro under different physiological and pathological conditions. Many of these targets are predicted with the help of bioinformatics software that may be suitable for miR-212, which has complicated the study of molecular targets of miR-212. Therefore, the use of high throughput tests (HTPs) such as micro-array studies and next generation sequencing (NGS) seems to be an appropriate solution for clarifying the effect of miR-212 on different molecular targets in cells (46), which was true for the study of miR-212 in this study, and the changes in the expression of miR-212 following infection of skin cells with T. rubrum could be attributed to this issue.
The level of EGFR gene's expression also changed in skin cells after infection with T. rubrum. In fungal lesions, the expression of EGFR gene was reduced, and in the healthy skin around the lesion, the expression of this gene was enhanced. The simultaneous changes of EGFR and miR-212 genes in Trichophyton-infected skin cells indicate an increase in the expression of miR-212 and a decline in the expression of EGFR, but these changes in the healthy skin cells around the fungal lesion involve a reduction in the expression of miR-212 and an elevation in the expression of EGFR.
5.1. Conclusions
Bioinformatics analysis showed that miR-212 could affect EGFR as a potential target. Thus, miR-212 may also target EGFR gene in infected cells with T. rubrum, which needs further practical studies. As shown in Diagram 1 in the findings, miR-212 in tissues infected with dermatophyte caused changes in infected tissues, such that T. rubrum significantly reduced the expression of EGFR gene and increased the miR-212 gene expression to eight times higher than EGFR gene expression, which may be the cause of dermatophytosis. On the other hand, in control tissue samples, the miR-212 gene expression was much lower than EGFR gene expression, which may indicate that the patient has been able to resist the onset of dermatophytic infections of T. rubrum in the marginal areas of the dermatophyte lesions caused by the EGFR gene.