1. Background
Cryptosporidium is a coccidian protozoan parasite, which belongs to the phylum Apicomplexa and causes cryptosporidiosis. The spread of infection hinges on several factors such as environmental situation, host growing old, immune system status, geographic locality, and contact with infected humans and/or animals (1). Epithelial cells contribute primary and quick defense in opposition to cryptosporidiosis, and also activate immune effector cells to the infectivity position. The attachment of Cryptosporidium parvum to apical cell surface can stimulate host cell signal pathways and hence modify cell function (2). The development of C. parvum infection in intestinal epithelial cells is effectively controlled by both type I and II interferons (IFNs) (3). Type I IFNs bring cell-inherent antimicrobial states in infected and adjacent cells that confine the increase of infectious agents, mainly viral pathogens.
The investigation on type I IFNs in immunity to parasitic protozoa is limited; however, the role of type I IFNs showed that they reduce the severity of INF of Leishmania major, and that they can act in a straight line on host cells to induce parasite inactivation. Some studies showed that the development of C. parvum infection in intestinal epithelial cells is effectively controlled both by type I and type II IFNs. The enterocytes can produce type I IFNs in rejoinder to C. parvum infection and lead to the inhibition of parasite development (3). The cultivation of C. parvum in tissue culture provided a new approach to recognize different gene expressions and immunological responses to different parasite proteins. Initially, primary epithelial cell culture (PECs) can stimulate different cellular genes after being exposed to C. parvum (4). Additionally, the continuous cell lines HCT8 and HT-29 showed good support for C. parvum (5). The implementation of advanced molecular tool can assess the gene expressions and their relative functions. The application of RT2Profiler™ PCR Array is a golden standard, which can measure low levels of gene expression (6).
2. Objectives
Since very few publications with focus on gene expression profile in the cells exposed to C. parvum oocysts can be found, the current study aimed at evaluating overexpressed and underexpressed genes related to type I INFs in HT29 cells infected by C. parvum using RT2 Profiler™ PCR Array.
3. Methods
3.1. Ethics Statement
The current study was granted by research deputy (Grant no: 94147) and ethically approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences.
3.2. Parasite Provision and Infection of Epithelial Cells
Cryptosporidium parvum oocysts (Iowa strain, BTF Company from Sydney –Australia) were kept in a solution of 0.01% Tween 20, 100 U/mL penicillin, and 100 mg/mL gentamicin, and stored at 4°C. To determine the infectivity of C. parvum, two-day-old neonatal rats (Razi Vaccine and Serum Research Institute, Karaj, Iran) were inoculated orally with 100,000 - 120,000 C. parvum oocysts (7). The shedding of the oocysts in the feces of neonatal rats were observed after eight days following the infection and confirmed with Ziehl–Neelsen staining.
3.3. Treatment of Oocysts
Oocysts were washed with 0.15 M/L phosphate-buffered saline (PBS; pH 7.2), centrifuged (3000 g for 15 minutes at 4°C) and treated with acidic H2O (pH 2.5; 20 minutes, vortexed at five minutes interval at 37°C). The oocysts were centrifuged (3000 g for four minutes at 20°C), re-suspended in parasite-maintenance Dulbecco's modified eagle's medium (DMEM; Gibco, Auckland, New Zealand) containing 4.5 g/L d-glucose, 0.58 g/L l-glutamine, 3.7 g/L sodium bicarbonate, 0.20 g/L bovine bile, 0.004 g/L folic acid, 0.001 g/L 4-aminobenzoic acid, 0.004 g/L d-calcium pantothenate, 0.88 g/L ascorbic acid, 1% heat-inactivated FBS, 2.4 g/L HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 100,000 U/L penicillin, 100,000 mg/L streptomycin, and 250 mg/L amphotericin B, adjusted to pH 7.4), and incubated for three hours at 37°C. The supernatant was removed, fresh DMEM was added, and oocysts were kept at 37°C for 90 minutes. The final dose of oocysts was adjusted 5 000/well prior to inoculation and cell culture (8).
3.4. In Vitro Cultivation
3.4.1. Preparation and Infection of Host Cells
The cell line HT-29 was purchased from Iranian Biological Research Centre, Tehran. The HT-29 cells were harvested in 25-cm2 flasks, followed by subculture in 24-well plates, supplemented with DMEM containing 10% fatal bovine serum (FBS; Gibco, Auckland, New Zealand), 100 U/mL penicillin, 100µg/mL streptomycin, and 25 µg/mL amphotericin B incubated with 5% CO2 at 37°C for 48 hours, and confluent monolayer cells were developed. For 24-well plates, the supernatant DMEM of the wells were removed and washed once with PBS (pH = 7.2). Next, 100 µL DMEM containing 5000 oocysts were inoculated onto each well, and the plates were incubated with 5% CO2 at 37°C for six hours. The 24-well plates were also inoculated with 100 µL DMEM containing 5000 oocysts/cm2 and incubated with 5% CO2 at 37°C for 24 hours. A 24-well plate was inoculated with PBS as control (mock). The HT-29 cell lines were exposed to C. parvum for 6 and 24 hours, and then, the cells were lysed with buffer RLT (RNeasy mini kit, Qiagen).
3.5. RNA Isolation and RT2 Profiler™ PCR Array
The total RNA was extracted for 6 and 24 hours separately using RNeasy mini kit (Qiagen), as per the manufacturer’s instructions. The cDNA was prepared using RT2 first standard kit (Qiagen). The qPCR Array was performed using the RT2 Profiler™ PCR Array of SABiosciences (http://www.sabiosciences.com/ArrayList.php) according to the manufacturer’s instruction. The triplicate test was conducted on each sample. Briefly, 2700 µL reaction mixture containing 1350 µL 2x RT2 SYBR Green qPCR Mastermix, 102µL cDNA, and 1248 µL RNAase-free water were prepared. The 25 μL of reaction mixture was added to each 96-well custom RT2 Profiler™ PCR Arrays and was subjected to real-time PCR (Applied Biosystems, Foster City, CA) with the following program: 95°C, 10 minutes, one cycle, 40 cycle, 15 seconds, 95°C, followed by 60°C for one minute (fluorescence data collection). The threshold cycle (Ct) for each well was calculated as described in web-based PCR Array Data Analysis Software (www.SABiosciences.com/pcrarraydataanalysis.php). A triplicate test was conducted to evaluate gene expression.
The gene expressions of the each sample were appraised on 84 genes (Table 1) related to human type I INFs (PAHS-016ZA). In addition, five housekeeping genes (ACTB, B2M, GAPDH, HPRT1, PRLPO), one genomic DNA control (GDC), three reversed transcription control (RTC), and three positive PCR control were used in the assay. The following overexpression and underexpression values were determined by fold change assessment. Fold-change (2^ [-delta delta Ct]) is the normalized gene expression (2^(-delta Ct)) in the test sample divided by the normalized gene expression (2^ [-delta Ct]) in the control sample. Fold-regulation represents fold-change results in a biologically significant way. Fold-change values > 1 indicate a positive or upregulation; the fold-regulation is equal to the fold-change. Fold-change values > 1 indicate a negative or down-regulation, and the fold-regulation is the negative inverse of the fold-change.
| Gene Symbol | Description |
|---|---|
| ADAR | Adenosine deaminase, RNA-specific |
| BAG3 | BCL2-associated athanogene 3 |
| BST2 | Bone marrow stromal cell antigen 2 |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| CAV1 | Caveolin 1, caveolae protein, 22 kDa |
| CCL2 | Chemokine (C-C motif) ligand 2 |
| CCL5 | Chemokine (C-C motif) ligand 5 |
| CD70 | CD70 molecule |
| CD80 | CD80 molecule |
| CD86 | CD86 molecule |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| CIITA | Class II, major histocompatibility complex, transactivator |
| CRP | C-reactive protein, pentraxin-related |
| CXCL10 | Chemokine(C-X-C motif) ligand 10 |
| DDX58 | DEAD(Asp-Glu-Ala-Asp) box polypeptide 58 |
| EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 |
| GBP1 | Guanylate binding protein 1, interferon-inducible |
| HLA-A | Major histocompatibility complex, class I, A |
| HLA-B | Major histocompatibility complex, class I, B |
| HLA-E | Major histocompatibility complex, class I, E |
| HLA-G | Major histocompatibility complex, class I, G |
| IFI16 | Interferon, gamma-inducible protein 16 |
| IFI27 | Interferon, alpha-inducible protein 27 |
| IFI30 | Interferon, gamma-inducible protein 30 |
| IFI6 | Interferon, alpha-inducible protein 6 |
| IFIH1 | Interferon induced with helicase C domain 1 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 |
| IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 |
| IFITM1 | Interferon induced transmembrane protein 1(9–27) |
| IFITM2 | Interferon induced transmembrane protein 2(1–8D) |
| IFI | Interferon induced transmembrane protein 3 |
| IFNA1 | Interferon, alpha 1 |
| IFNA2 | Interferon, alpha 2 |
| IFNA4 | Interferon, alpha 4 |
| IFNAR1 | Interferon (alpha, beta and omega) receptor 1 |
| IFNAR2 | Interferon (alpha, beta and omega) receptor 2 |
| IFNB1 | Interferon, beta 1, fibroblast |
| IFNE | Interferon, epsilon |
| IFNW1 | Interferon, omega 1 |
| IL10 | Interleukin 10 |
| IL15 | Interleukin 15 |
| IL6 | Interleukin 6 (interferon, beta 2) |
| IRF1 | Interferon regulatory factor 1 |
| IRF2 | Interferon regulatory factor 2 |
| IRF3 | Interferon regulatory factor 3 |
| IRF5 | Interferon regulatory factor 5 |
| IRF7 | Interferon regulatory factor 7 |
| IRF9 | Interferon regulatory factor 9 |
| ISG15 | ubiquitin-like modifier |
| ISG20 | Interferon stimulated exonuclease gene 20kDa |
| JAK1 | Janus kinase 1 |
| JAK2 | Janus kinase 2 |
| MAL | Mal, T-cell differentiation protein |
| MET | Met proto-oncogene(hepatocyte growth factor receptor) |
| MNDA | Myeloid cell nuclear differentiation antigen |
| MX1 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78(mouse) |
| MX2 | Myxovirus (influenza virus) resistance 2(mouse) |
| MYD88 | Myeloid differentiation primary response gene(88) |
| NMI | N-myc (and STAT) interactor NM_ NOS2 Nitric oxide synthase 2, inducible |
| OAS1 | 2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| OAS2 | 2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| PML | Promyelocytic leukemia |
| PRKCZ | Protein kinase C, zeta |
| PSME2 | Proteasome(prosome, macropain) activator subunit 2(PA28 beta) |
| SH2D1A | SH2 domain containing 1A |
| SHB | Src homology 2 domain containing adaptor protein B |
| SOCS1 | Suppressor of cytokine signaling 1 |
| STAT1 | Signal transducer and activator of transcription 1, 91kDa |
| STAT2 | Signal transducer and activator of transcription 2, 113kDa |
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) |
| TAP1 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| TICAM1 | Toll-like receptor adaptor molecule 1 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 |
| TLR3 | Toll-like receptor 3 |
| TLR7 | Toll-like receptor 7 |
| TLR8 | Toll-like receptor 8 Toll-like receptor |
| TMEM173 | Transmembrane protein 173 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 |
| TRAF3 | TNF receptor-associated factor 3 |
| TYK2 | Tyrosine kinase 2 |
| B2M | Beta-2-microglobulin |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| RPLP0 | Ribosomal protein, large, P0 |
| HGDC | Human genomic DNA contamination |
| RTC | Reverse transcription control |
| RTC | Reverse transcription control |
| RTC | Reverse transcription control |
| PPC | Positive PCR control |
| PPC | Positive PCR control |
| PPC | Positive PCR control |
3.6. Statistical Analysis
The results of overexpression and underexpression of each gene related to type I INFs were compared with mock cells using online Software (www.SABiosciences.com/pcrarraydataanalysis.php). In vitro, cellular assays were done in triplicate. Data were analyzed using student t test. Based on fold change, the overexpression of genes and underexpression of genes were determined for six and 24 hours after infection. P value < 0.05 was considered significant. The P values are calculated based on a Student t test of the replicate 2^(-delta Ct) values for each gene in the control and treatment groups. The automatic selection from housekeeping genes with genes (HKG) panel with ACTB and GAPDH was conducted to evaluate the normalization of the test samples for six and 24 hours. This method automatically selects an optimal set of internal control/housekeeping/normalization genes for the analysis from the available HKG panel on the PCR array.
4. Results
4.1. Inoculate Cryptosporidium parvum Iowa Strain
The post-infected neonatal rat with C. parvum showed symptoms such as weakness, severe diarrhea, dehydration, and volume depletion three to four days after oral inoculation. The microscopy presence of Cryptosporidium oocysts in feces of the infected neonatal rats was stained by Ziehl-Neelsen. The amount of 522 ng/µL of the extracted RNA from exposed cells to C. parvum was used for RT² PCR™ Array.
4.2. Gene Expression Pattern of Human Type I INF Response at RT² Profiler™ PCR Array
Based on technical triplicate data, fold change for the significant upregulation of the expressed gene was defined (fold-change > 1.0: P < 0.05) for six and 24 hours and fold change for significant downregulated gene was described (fold-change < 1.0; P < 0.05) following the infection. It should be noted that the GSEA analysis confirmed all the overexpressed genes related to type I INFs pathway.
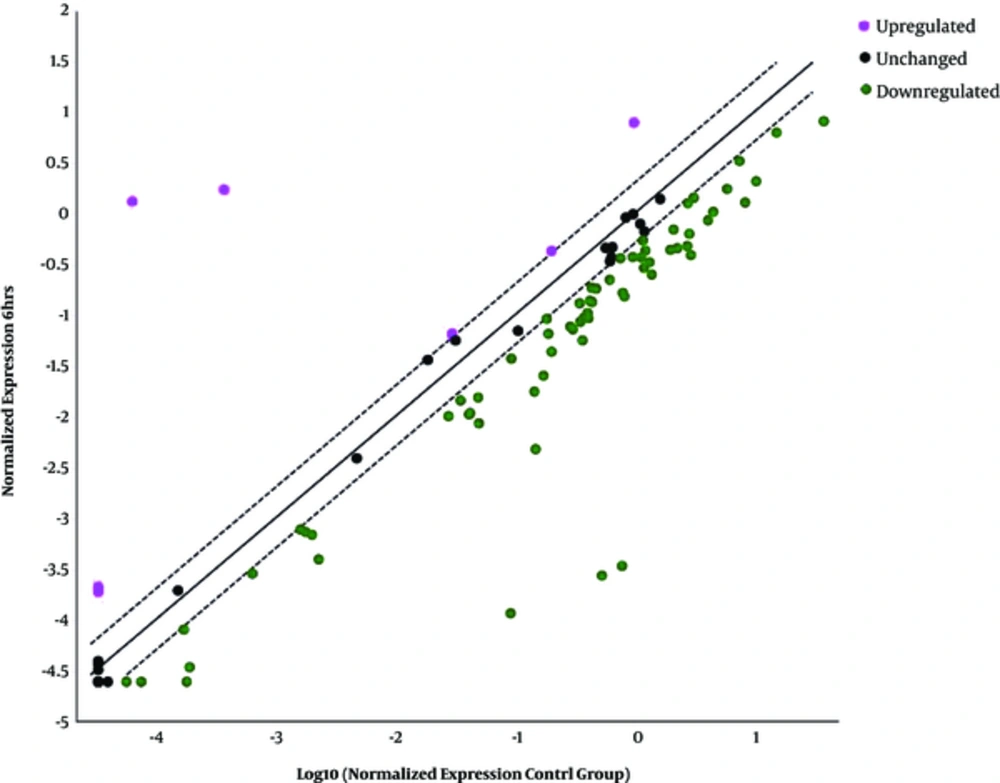
4.3. Overexpressed, Underexpressed, and Unchanged Genes Six Hours Post-Infection
Four top overexpressed genes related to type I INFs namely HLA-A, IL-10, MX1, and SH2D1A were selected (Table 2 and Figure 1) and out of 60 downexpressed genes, five genes including IRF2, IRF5, PML, TIMP1, and TYK2 were chosen for further investigation (Table 3 and Figure 1). The remaining 55 underexpressed genes were as follows: CAV1, TAP1, TLR9, CD80, JAK1, IFI30, IFI27, IFITM2, SHB, OAS2, IFI6, IFNAR2, BST2, ADAR, STAT2, MET, PRKCZ, IRF9, IFNAR1, MX2, TICAM1, SOCS1, JAK2, IFI16, TRAF3, STAT1, BAG3, TLR3, ISG15, TNFSF10, IRF3, IFNA2, HLA-G, CXCL10, ISG20, STAT3, CASP1, IRF7, MYD88, NMI, IL15, IFITM3, IFNE, IFNA1, IRF1, IFITM1, IFNW1, IFIT1, IL6, IFIT3, IFIH1, NOS2, IFIT2, CIITA, and CCL5. Twenty genes residual unchanged were as follows: CCL2, CD70, CD86, CDKN1B, CRP, DDX58, EIF2AK2, GBP1, HLA-B, HLA-E, IFNA4, IFNB1, MAL, MNDA, OAS1, PSME2, TLR7, TLR8, TMEM173, and VEGFA.
| Overexpressed Genes | Fold-change 6 h | P-value h |
|---|---|---|
| HLA-A | 4614.27 | 0.019548 |
| IL10 | 6.43 | 0.031975 |
| MX1 | 20709.49 | 0.019887 |
| SH2D1A | 5.74 | 0.003034 |
| Under expressed Genes | Fold change 6 hours | P value hours |
|---|---|---|
| IRF2 | -31.44 | 0.037850 |
| IRF5 | -801.25 | 0.028377 |
| PML | -1977.58 | 0.001647 |
| TIMP1 | -2343.40 | 0.008456 |
| TYK2 | -8.34 | 0.042893 |
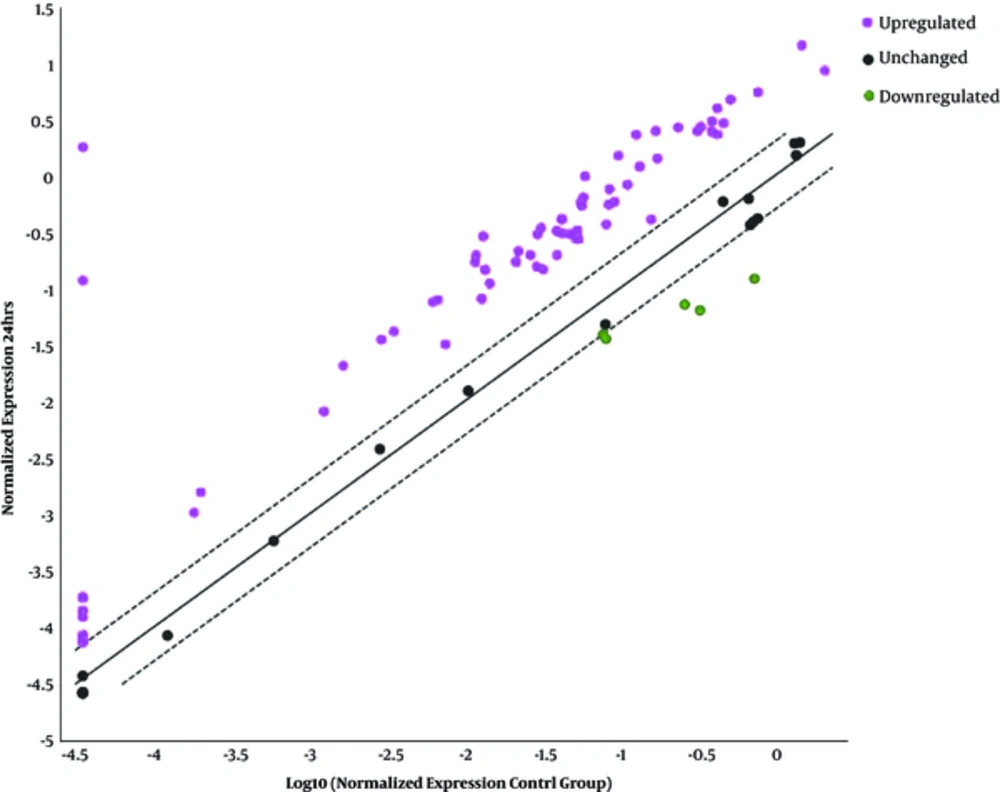
4.4. Overexpressed, Underexpressed, and Unchanged Genes 24 Hours Post-Infection
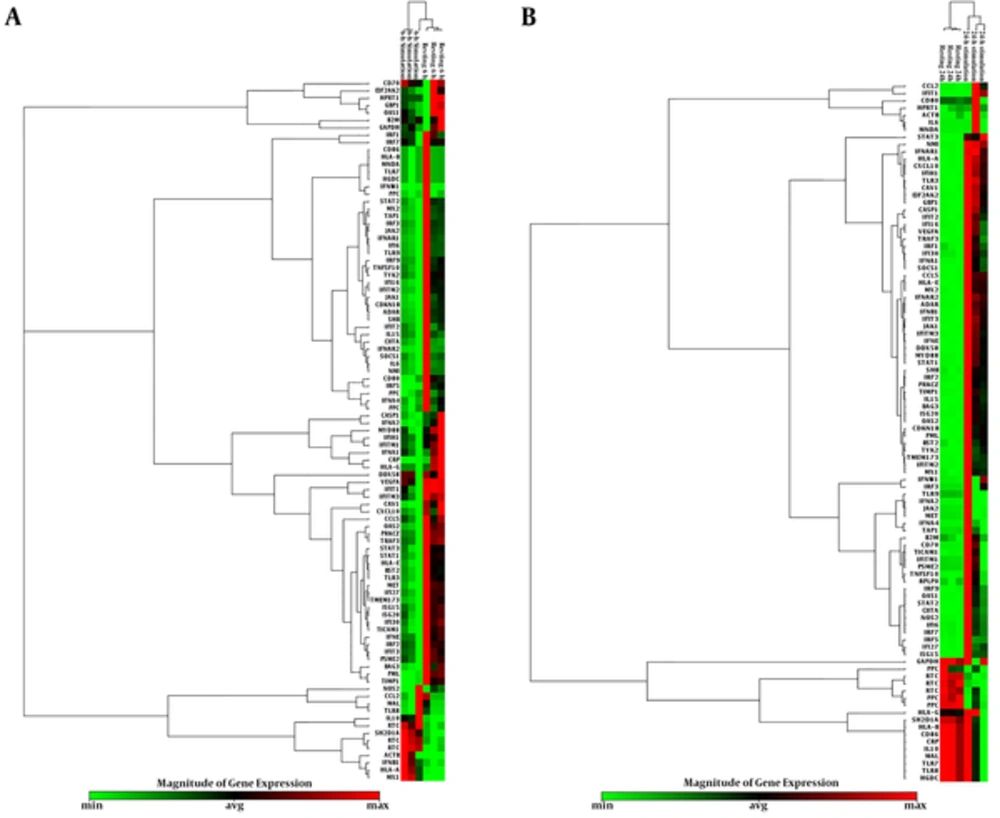
Sixty-seven genes were overexpressed among which 10 top genes such as CASP1, CXCL10, DDX58, IFIH1, IFNAR2, IL15, MYD88, NMI, STAT3, and TLR3 were selected for further studies (Table 4, Figure 2). The following 57 overexpressed genes were as follows: IFNE, IFNB1, CAV1, EIF2AK2, CDKN1B, ISG15, CCL5, ISG20, BAG3, PRKCZ, GBP1, IFNAR1, IFITM3, TIMP1, STAT1, HLA-A, VEGFA, JAK1, IFIT2, IFIT3, ADAR, TRAF3, IFNA1, OAS2, HLA-E, TMEM173, IRF2, IFITM2, IFI16, IFI27, SOCS1, MX2, PML, TYK2, SHB, OAS1, MX1, IFI30, CIITA, BST2, NOS2, IRF7, IRF1, CD70, TNFSF10, STAT2, TICAM1, IRF9, IRF5, IFNW1, IFI6, IFNA4, TAP1, CCL2, MNDA, IFNA2, and IL6. Three underexpressed genes IFITM1, PSME2, and IRF3 were observed in post-infected cells. The remaining 14 unchanged genes related to type I INFs were diagnosed in the infected cells as follows: TLR7, TLR8, TLR9, SH2D1A, JAK2, MAL, MET, IL10, IFIT1, HLA-B, HLA-G, CRP, CD80, and CD86. The cluster gram pattern showed that the overexpressed and underexpressed genes were related to human type I INF after six and 24 hours exposure (Figure 3 A & B).
| Overexpressed Gene | Fold-change 24 h | P-value 24 h |
|---|---|---|
| CASP1 | 12.46 | 0.008962 |
| CXCL10 | 21.76 | 0.002424 |
| DDX58 | 50719.61 | 0.008805 |
| IFIH1 | 14.42 | 0.002223 |
| IFNAR2 | 14.50 | 0.002332 |
| IL15 | 16.48 | 0.022768 |
| MYD88 | 16.37 | 0.009969 |
| NMI | 17.81 | 0.000123 |
| STAT3 | 15.13 | 0.011018 |
| TLR3 | 12.05 | 0.002110 |
5. Discussion
It is important to understand whether the reaction of exposed cells to C. parvum cells are resistant or the disease develops. Therefore, it is important to evaluate the innate immune genes related to type I INF response. Limited information is available to describe immune response expression of genes in cells exposed to C. parvum. Therefore, the interest of the current study was to find out overexpression and underexpression of related genes to human type I INFs (α and β) in HT-29 cells exposed to C. parvum for six and 24 hours. Based on fold-change outcomes, four top overexpressed genes with fold-change > 1.0 (P < 0.05) including HLA-A and MX1 genes were evaluated in infected cells for six hours. Birte Pantenburg et al., reported that the HLA-A and -B were involved in presenting the C. parvum antigens to CD8 + T-lymphocytes to eliminate the parasite (8). The current study found that the HLA-A gene was overexpressed in infected cells after six hours exposure. This finding was in agreement with what Birte Pantenburg et al., reported.
IFN-α and -β induce MX1 gene expression that encodes a GTPase activity and acts as a potent inhibitor for replication of many RNA viruses and Salmonella typhimurium (9-11). The current study data showed that MX1 gene was over expressed in the exposed cell six hours post-exposure. It was thought-provoking to express the MX1 gene as an antiviral factor induced by a C. parvum protein, which needs further investigation. Expression of SH2D1A gene synthesizes protein 2B4, which can interact with SLAM (signaling lymphocyte-activating molecule) to monitor T-cell and B-cell response to Epstein-Barr virus (EBV) infection (12-14). The current study findings revealed that SH2D1A gene was overexpressed in vitro for six hours. It is necessary to investigate the characterization of antigen or protein that induces SH2D1A gene expression against C. parvum infection. Wyatt et al., found that C. parvum epitopes (44 to > 200 kD) could prompt overexpression of IL-10 gene in intestinal epithelial cells. They concluded that the expression of IL-10 gene may have an important role in developing parasite infection (15). Based on the current study data, IL-10 gene was overexpressed in the infected cells.
The following genes, TIMP1, TYK2, IRF2, PML, and IRF5 were under expressed in the infected cells for six hours. The expression of TIMP1 (tissue inhibitors of metalloproteinases) gene inhibits expression of MMPs (matrix metalloproteinases) gene, which is implicated in developing cancer, neurological, and infectious pathologies (16). Overexpression of TIMP1 gene is implicated in clearance of parasites infection (16, 17). According to the current study findings, the TIMP1 gene was underexpressed for six hours, but overexpressed in the infected cells for 24 hours. Thus, it was concluded that the overexpression of TIMP1 and SH2D1A genes might be an important key for C. parvum to become self-limited infection.
The expression of TYK2 gene plays a significant role in the intracellular signaling of type I IFNs using phosphorylation and activation of STAT proteins (18). The expression of TYK2 gene plays a vital role in the synthesis of INF-β. The stimulation of INF-β production via overexpression of TYK2 gene is detected in the infected cells by L. major. The current study observed that TYK2 gene was downexpressed for six hours, but overexpressed for 24 hours following the infection. In contrast, Piyal Ganguli stated that leishmaniosis stimulated the production of INF-β, which in turn can upregulate the secretion of RAP1 and SOCS3 proteins within the T-cells that potentially inhibits MAPK and JAK-STAT signaling pathway via TYK2 mediated pathway (19).
The expression of IRF2 (interferon regulatory factor 2) gene upregulated type I INF associated with signaling pathway and resulted in the synthesis of IL-12 in cells infected by L. major (20, 21). IRF2 protein acts multiple functions such as transcription factor, IRF2, transcriptional activator of histone H4, IFN-stimulated regulatory element (ISRE)-responsive genes and requires optimal maturation of natural killer (NK) cells (22). The current study recorded the IRF2 gene as underexpressed for six hours, but it was overexpressed for 24 hours. Thus, it is very important to investigate the characterization of a protein of C. parvum to induce IRF2 gene expression.
Promyelocytic leukemia (PML) gene produces a protein, acts as a tumour suppress and prevents cells from growing and dividing. Promyelocytic leukemia protein is also a cofactor to induce apoptosis. In the current study, the PML gene was downexpressed in the infected cells for six and 24 hours. TLR 7, TLR 9 via signaling pathway of INF type I induced IRF5 gene expression, which activates pro-inflammatory cytokines such as IL6 and TNF (tumor necrosis factor) (23, 24). The expression of IRF5 gene was detected in the infected cells by L. donovani (25). The current study data showed that IRF5 gene was downexpressed in the infected cells for six and 24 hours. The current study selected 10 top overexpressed genes with high enrichment including IL15, DDX58, CXCL10, NMI, MYD88, STAT3, IFNAR2, IFIH1, CASP1, and TLR3 genes relevant to types I IFNs in post-infected cells for 24 hours. IL-15 (mRNA is overexpressed in cells of secretary tissues, dendritic cells (DCs), epithelial cells, bone marrow stromal cells, and fibroblasts) and its functions stimulate the proliferation of T-lymphocytes and important role of IL-15 to control infection (26-28). The current study results indicated that IL-15 gene was overexpressed in the infected cell for 24 hours; to the authors` best knowledge, an overexpression of IL-15 is reported in infected cells by Cryptosporidiosis.
The expression of DDX58 gene can promote immunological responses (29). Alvarez et al., reported that DDX58 gene was downexpressed in lamb liver tissue cells infected by Fasiola hepatica (30). In contrast, the current study found that DDX58 gene was overexpressed in the infected cells for 24 hours. CXCL10 is a Chemokine and can attract the immune cells at the site of infection, while in the patients with AIDS, due to lack of immune cells, CXCL10 is enabled to recruit immune inflammatory cells causing immune pathogenesis. Carneiro et al., reported that the genes related to IFN-γ such as CXCL10 were upregulated when HPs and LPs (high or low IFN-γ producers) following motivation of peripheral blood cells were exposed to L. braziliense, which was in line with the current study findings (31). The expression of CXCL10 gene via stimulation of IFN-δ gene was detected in intestinal epithelial cells infected by C. parvum (32). Type I IFN and IFN-γ stimulated the expression of N-myc interactor (Nmi) gene (33). NMI is a cytokine with a multiple function including enhancement IFN-γ-induced STAT-dependent transcription and increased interleukin-2 (IL-2)-driven STAT5-dependent relative transcription (34). NMI protein acts as antiviral activities against prototype foamy virus (35), Sendai virus (33), and foot and mouth disease virus (FMDV) (36). To the authors` best knowledge, parasite induced NMI protein is not reported yet. In the current study, the NMI gene was overexpressed in the infected cells for 24 hours.
MyD88 gene encodes a protein and implicates in innate immunity. Toll-like receptor 4 induced type I INF via MYD88 (37). Lacroix-Lamande et al., injected polyC to neonatal mice infected with C. parvum, and found enhancement signalling MYD88 pathway, which resulted in resistance of mice intestine cells to parasite oocysts infection (38, 39). Rogers et al., reported that mice lacking MyD88 were more at risk to infection than the wild-type mice (38). In the current study, MYD88 gene was overexpressed for 24 hours indicating an important key role in activation of type I INF. The TLR3 protein performs multiple functions such as pathogen recognition, activation of innate immunity, recognition of dsRNA associated with viral infection, and inducing NF-Кβ and type I INF production (40). The current study observed that the TLR-3 gene was overexpressed for 24 hours in vitro, which was in agreement with the findings of Koblansky et al., (41) but in contrast with those of the study by Beiting (42). STAT3 is a transcription factor that can be activated by interferon ligand that implicates in apoptosis and cell growth processes (43). Kyeong et al. showed that STAT3 gene was downexpressed in Caco-2 cells infected with Entamoeba histolytica, but it was in contrast with the current study results. The STAT3 gene was overexpressed for 24 hours (44).
Caspase-1, which can activate two inflammatory cytokines and initiate the pro-apoptosis program, results in cell death (45). Liu et al., reported that about 20% of infected cells led to apoptosis for 48 hours, but the majority of cells were protected from apoptosis by C. parvum infection. They found high levels of caspase-3, -4, -6, and -7 activation and low levels of caspase-2, -8, and -9 activation, while no caspase-1 and -10 genes were expressed at a 48-hour course of infection. Cryptosporidium parvum suppressed staurosporine-induced apoptosis and initiated caspase-3/7 activation of intestinal epithelial cells via upregulation of survivin, which sustains parasite infection (46). The current study found that the caspase-1 gene was overexpressed for 24 hours. In the current study, caspase-2-10 gene expressions were not evaluated, even though it was necessary for further study.
IFNAR2 (interferon-alpha/beta receptor beta) protein bands activate the receptor stimulation Janus protein kinases, which in turn activate STAT1 and STAT2. It subsequently produces type I IFN and pro-inflammatory cytokines, in that way up regulating a family of IFN-stimulated genes (ISGs) that exert pleiotropic inhibitory effects on viral reproduction in the neighboring cells (47, 48). The overexpression of IFNAR2 gene is yet to be reported in the infected cell by C. parvum. The current study found that the IFNAR2 gene was overexpressed in the infected cells for 24 hours, even though its functions should be investigated.
IFIH1 (interferon-induced helicase 1) recognizes dsRNA of many viruses and acts as mediator for antiviral response (49). The IFIH1 gene was overexpressed, but its function requires further evaluation. The cells infected by C. parvum result in overexpressed genes related to subtype 1 interferon including synthesis of MX1, SH2D1A, TLR3, NMI, and IFIH1 proteins, which may have antiviral properties and are promising new candidate biological drugs against some viral infections. Han et al., reported that anti-viral cytokine interferon-β (IFN-β) was produced after IMs (inflammatory monocytes) exposed to Toxoplasma gondii (50).
6. Conclusions
In conclusion, the overexpression of TIMP1 gene had an important role in the clearance of C. parvum. The overexpression of TIMP1, SH2D1A, and MYD88 genes may lead C. parvum to become a self-limited infection. The overexpressed TIMP1 may have anticancer properties. The overexpression of TYK2 and MYD88 genes synthetizing type I INF may play an important role in innate immunity. It is important to investigate the induction of production the protein and overexpression of IRF2 gene with multiple functions related to subtype I INF. The CXCL10 gene function is involved in the attraction of immune effector cells to the site of infection, the overexpressed CXCL10 was observed. The current study identified the upregulated genes, which can prevent the invasion of parasite to cells. Finally, the identification of all unknown induced overexpressed and underexpressed genes by C. parvum requires thorough investigation. In the light of aforementioned overexpressed genes related to type I IFN as a part of innate immunity may play an important role against C. parvum infection.