1. Background
Giardia duodenalis (G. duodenalis) is an enteric flagellated parasite, having several host species worldwide (1). This parasite can be transmitted by contamination though oral-fecal route (2, 3). Direct life cycle of G. duodenalis composed of 2 main stages: the infective cyst, resistant to environmental conditions, and the vegetative trophozoites, which live in the lumen of the small intestine and can cause a problem in infected individuals (4-6). Annually, about 200 million people are affected with symptomatic giardiasis in Asia, Africa, and Latin America (6, 7). Giardia infections cause an economic burden in public health and veterinary (3, 8-10). Nowadays this protozoan parasite is considered as one of the most common causes of diarrhea in humans. In children, association between giardiasis and malnutrition, growth reduction, and cognitive disorders have been commonly seen (6, 11-13).
The successful treatment and control strategies of G. duodenalis require proper and rapid techniques for accurate identification of this infectious micro-organism. Several laboratory techniques with different sensitivities are commonly used for detection of giardiasis, such as traditional microscopy method, enzyme-linked immunosorbent assay (ELISA), and direct fluorescent antibody test (DFA) (4, 5, 10, 14-16). However, microscopic examination is characterized as a definitive method for the diagnosis of giardiasis, however, it has some drawbacks such as expertise requirement in identifying the micro-organism and intermittent cyst shedding of parasite in stool (3, 8, 10). The molecular diagnostic techniques used as rapid and powerful tools for detection of G. duodenalis in environmental and clinical specimens and have enhanced our knowledge about the taxonomy, epidemiology, and molecular diversity of this parasite (3, 17-21).
Molecular PCR based assays have been successfully applied for characterization of Giardia genotypes using the small subunit ribosomal RNA (SSU rRNA); beta-giardin (bg); elongation factor 1-α; glutamate dehydrogenase (gdh) and triose phosphate isomerase (tpi) genes, that are extremely different in genetic variability (1, 5, 22-27). Among these markers the partial gdh, bg, tpi, and SSU rRNA genes are mostly used for identification of this parasite at the assemblage level (6, 28). In order to achieve a highly sensitive PCR amplification as a consistent and reliable method in detecting low amount of target DNA, an effective extraction method should be carefully selected and established (4, 8, 29, 30). In molecular techniques, PCR inhibitors are among the most important factors leading to low sensitivity or failure in PCR amplification. These inhibitors consist of all existing substances in fecal specimens that have been classified into different types, according to their sources such as lipids, hemoglobin, polysaccharides and bile salts. In addition, other compounds are remaining during the process of cyst separation, DNA extraction, and purification steps. In DNA extraction of Giardia, the next major challenge in addition to inhibitors is the efficient break down of the cyst wall (21, 30).
In order to reduce these complications some essential and useful handling approaches such as proper sample preparation, separation of cyst and disruption of the wall, removal or reduction of inhibitors during extraction or purification of target DNA, and finally use of effective compounds for elimination of the PCR inhibitors should be carefully considered. For decreasing the fecal substances, isolation and purification of G. duodenalis cysts from feces using correct concentration techniques such as sucrose flotation should be considered as one of the useful preparatory procedures prior to DNA extraction. Another helpful approach to consider is the numerous cycles of freeze-thawing and adding glass bead before the nucleic acid extraction, which facilitate the breakdown of the cyst wall and consequently lead to increase in the quantity of the extracted DNA (4, 29). In addition, a variety of compounds have been applied for removal of inhibitors in PCR amplification, such as bovine serum albumin (BSA), which seem to exert a positive influence (21, 31).
Various numbers of in-house methods such as the traditional Phenol-Chloroform Isoamyl alcohol method and commercial kits like QIAamp DNA stool minikit (QIAGEN, Germany) have been assessed (4, 5, 8, 22, 29, 30) for DNA extraction of G. duodenalis from stool specimens. These protocols, which are based on mechanical, chemical, and/or enzymatic lyses have been enabled researchers to extract, purify, and subsequently evaluate DNA of Giardia in molecular analysis. These techniques have advantages over the others due to cost-effectiveness, speed in processing, simplicity, and low limitations (4, 8, 29, 30, 32-35).
2. Objectives
The objective of this study is to compare the quantity and purity of extracted DNA using 3 different DNA extraction techniques including the conventional Phenol-Chloroform Isoamyl alcohol method and the 2 commercial kits, QIAamp DNA stool mini kit (QIAGEN, Germany) and YTA Stool DNA Isolation mini Kit (Yekta tajhiz Azma, Iran) and to assess the potency of DNA in PCR amplification of a specific area of SSU rRNA gene.
3. Methods
3.1. Ethics Statement
The ethics committee of Ahvaz Jundishapur University of Medical Sciences approved this study (IR.AJUMS.Rec.OG-94150).
3.2. Sample Collection and Preparation
A total of 60 fecal specimens containing G. duodenalis cysts were taken from infected humans, referred to a laboratory of governmental hospital in city of Ahvaz, Southwest of Iran. All samples were labeled with numbers and transferred to the department of parasitology, school of Medicine, Ahvaz Jundishapur University of Medical Sciences and stored at 4°C without using any preservatives. A total of 20 positive samples with > 5 cysts in a field of 400 × magnification were selected using the direct microscopic analysis of wet mount smears. The samples were homogenized in order to get the regular number of cysts in prepared suspensions by diluting about 10 gram of each fecal specimen in 30 mL of distilled water. Then, suspensions were passed from 4 layer of gas, centrifuged at 600 × g for 5 minutes and the pellets were diluted, washed, and centrifuged in 2 repetitions. Purification of cysts was carried out by the sucrose flotation technique (28) with some modification, briefly using 30 mL of suspended fecal samples in distilled water, which were added to 15 mL of cooled sucrose solution (1 M) and centrifuged at 800 ×g for 5 minutes. The middle layer was then gently collected and washed for 3 consecutive times by adding distilled water and centrifuging at 600 × g for 5 minutes. Finally, the pellet was suspended in distilled water to a final volume of 1 mL and stored at -20°C for further DNA extraction.
3.3. DNA Extraction Techniques
In attempt to facilitate the breakdown of the cyst wall, the purified cysts were treated with 7 consecutive times of freeze-thawing consisting of freezing in liquid nitrogen for 5 minutes and boiling in water bath for 7 minutes. Subsequently, each treated sample were aliquoted in equal final volumes of 200 µL, in 3 separate microtubes and used for further DNA extraction using 3 above-mentioned methods. A NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, USA) was used in order to quantify and assess the purity of extracted DNA obtained from each method. The absorbance at 260 nm wavelength was taken in order to calculate DNA concentration, and the purity of DNA was assessed by the ratios of absorbance at 260/280 and 260/230. In this study, three different methods of DNA extraction (QIAamp DNA stool mini kit (QIAGEN, Germany), YTA Stool DNA Isolation mini Kit (Yekta tajhiz Azma, Iran), and Phenol-Chloroform Isoamyl alcohol) were examined and analyzed.
3.3.1. QIAamp DNA Stool Mini Kit Method
This commercial kit was used in order to extract genomic DNA using 200 µL of each aliquoted stool sample according to the manufacturer’s instructions with minor modifications. In the first step lysis buffer ASL was added to the fecal sample in 2 mL microtube and thoroughly homogenized by vortexing, followed by incubation at 95°C in water bath with the modification of increasing time to 15 minutes. After centrifugation, supernatant was transferred to a new microtube and InhibitEX tablet was added in order to remove the fecal contaminants and PCR inhibitors. Suspension was then centrifuged and supernatant incubated with proteinase K and buffer AL for 10 minutes at 70°C for enzymatic digestion and release of DNA. Ethanol absolute was added to the lysate from the previous step, mixed and was carefully passed through the silica membrane (QIAamp spin column) for binding DNA. In order to obtain pure DNA and eliminate inhibitory agents and contaminants, 2 washing steps were carried out using buffers AW1 and AW2. Finally extracted DNA was eluted in 100 µL buffer AE and stored at -20°C for further use.
3.3.2. Phenol-Chloroform Isoamyl Alcohol Method (PCI)
This method has been used for several types of biological samples such as blood, tissue, stool, and cultured cells. For DNA extraction using standard PCI protocol (4, 36) with some modifications, initially enzymatic digestion was performed by adding lysis buffer (Tris-HCL, 100 mM; EDTA, 0.5 M; SDS, 10%; proteinase K, 20 mg/mL) to 200 µL of aliquoted stool specimen followed by shaking and incubating for 3 hours at 56°C. In order to eliminate the contaminants and inhibitors, cetyltrimethylammonium bromide (CTAB) was added and homogenized sample was incubated at 65°C for 15 minutes.
In order to further remove protein contaminants and PCR inhibitors, the lysate was treated with Phenol/chloroform/Isoamyl alcohol (25:24:1) (Sigma-Aldrich Chemie GmbH). Then ice-cold 100% ethanol was added to the removed aqueous phase, gently mixed, and placed at -20°C for 24 hours in order to precipitate the extracted DNA. After centrifugation, supernatant was discarded and finally contaminants and PCR inhibitor compounds were washed twice with 75% ethanol. DNA was eluted by dissolving the dried DNA pellet in 50 µL sterile distilled water and was stored at -20°C until further use in molecular assays.
3.3.3. YTA Stool DNA Isolation Mini Kit Method
This commercial kit was designed for DNA extraction from fecal samples based on mechanical disruption with glass beads and enzymatic digestion, using proteinase K. In this study, all the procedures were carried out according to the recommendations of manufacturer’s instructions. Briefly 200 µL aliquoted fecal sample were thoroughly homogenized by adding 200 mg glass beads, buffer SDE1, as well as proteinase K and incubated for 20 minutes at 60°C. The process of lysis was then completed by using buffers SDE2, SDE3 and SDE4, respectively. After adding ethanol absolute, the lysate was transferred to SDE column in order to bind the released DNA to silica membrane. In order to eliminate PCR inhibitors and contaminants, washing buffer was further used for 2 consecutive times and purified DNA was eluted in 100 µl of preheated elution buffer and stored at -20°C before further use for PCR amplification.
3.4. Molecular Identification of G. Duodenalis
The efficiency of each extraction method for detection of this parasite was investigated by PCR amplification, targeting a 350-bp fragment of the small subunit ribosomal RNA (SSU rRNA) gene using primers GiF (5’-AGCCGGACACCGCTGGCAACC-3’) and GiR (5’-CGGCTGCTGGCACCAGACCTT-3’) as described previously (37), with minor modifications. The PCR was performed in the final volume of 25 µL reaction consisting of 5 µl of DNA template, 1 µL of each primer (Metabion, Germany) at 25 pmol concentration, 5.5 µL of deionized distilled water, and 12.5 µL Taq DNA Polymerase 2x Master Mix (AMPLIQON, Denmark) comprising of dNTP, MgCl2 (2 mM), Taq polymerase enzyme, tracking dye, and reaction buffer. PCR amplification was performed on a thermal cycler (Bio-Rad, My Cycler, US) using the following conditions: after initial denaturation at 94°C for 5 minutes, a set of 30 cycles comprising of denaturation at 94°C for 1 minutes, annealing at 67°C for 1 minutes, and extension at 72°C for 1 minutes followed by final extension at 72°C for 5 minutes. In order to validate the results, a Giardia-positive DNA sample and distilled water were included in all the PCR amplifications as positive and negative control, respectively. Finally products of each round of PCR were assessed by electrophoresis on 2% agarose (Merck, Germany) gels, stained with ethidium bromide and visualized using the UV transilluminator (Syngene, G: Box, UK).
3.5. Statistical Analysis
Results of DNA extraction and PCR amplification were analyzed using SPSS version 22 software. The assessment of diagnostic sensitivity of 3 different DNA extraction methods was carried out by the Chi-square test with probability levels (P value) < 0.05. In order to estimate statistical correlation of the measured values of DNA concentration, the ratios of A260/280 and A260/230 with DNA extraction method, the Kruscal-Wallis, and Mann-Whitney U test as non-parametric tests (based on Mean Rank) were performed.
4. Results
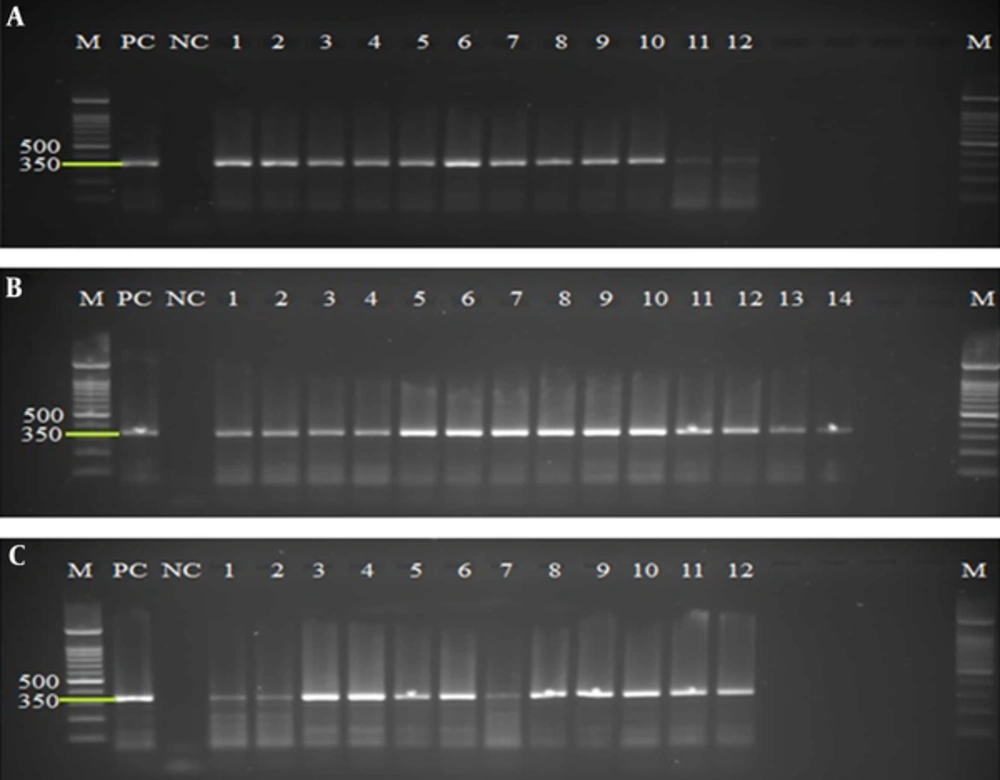
All extracted and purified DNA from concentrated and pretreated cysts of 20 fecal samples were evaluated, using amplification of a 350-bp fragment of SSU rRNA gene. The PCR results using DNA extracted from QIAamp DNA stool mini kit, phenol/chloroform/Isoamyl alcohol, and YTA Stool DNA Isolation mini Kit methods on agarose gel were represented successful amplification in 12, 14, and 12 samples, respectively (Figure 1).
A, PCR products of extracted DNA using QIAamp DNA stool mini kit protocol. B, PCR products of extracted DNA using the phenol/chloroform/Isoamyl alcohol protocol. C, PCR products of extracted DNA using the YTA Stool DNA Isolation mini Kit. Lines M, 100 bp molecular Marker (Cynagene, Iran); PC, Positive Control; NC, Negative Control; lines 1 - 14: Positive samples.
The Chi-square test was used for analyzing the efficiency of the DNA extraction method on the sensitivity of PCR diagnostic method. In this study QIAamp DNA stool mini kit, phenol/chloroform/Isoamyl alcohol and YTA Stool DNA Isolation mini Kit methods had a diagnostic sensitivity of 60%, 70%, and 60%, respectively (Table 1). Therefore, phenol/chloroform/Isoamyl alcohol was significantly (P < 0.05) demonstrated as the most effective method for obtaining the best PCR results. Comparison of the 3 different methods regarding DNA concentration and the ratios of A260/230 using Kruscal-Wallis and Mann-Whitney’s U-test has revealed statistically significant differences (α = 0.05, P < 0.0166). No significant differences were found in comparing the ratio of A260/280 using Kruscal-Wallis test (P > 0.05). Finally, the most concentrated DNA was obtained from phenol/chloroform/Isoamyl alcohol method and the best purity based on comparing the ratio of A260/230 was obtained from QIAamp DNA stool mini kit.
| Extraction Method | PCR | ||
|---|---|---|---|
| Positive | Negative | Diagnostic Sensitivity, % | |
| QIAamp DNA stool mini kit | 12 | 8 | 60 |
| Phenol/chloroform/Isoamyl alcohol | 14 | 6 | 70 |
| YTA Stool DNA Isolation mini Kit | 12 | 8 | 60 |
5. Discussion
Selection and careful implementation of an effective DNA extraction method with high quantity and quality of extracted DNA is considered as one of the most important steps in obtaining reliable and reproducible results in molecular assays. In addition, the ease-of-use, simplicity, and cost-effectiveness are among the essential selection criteria. However in some molecular assays, false-negative results have been reported despite the microscopic detection of G. duodenalis in wet mount smears of fecal specimens (4, 7, 28). The failure of target gene amplification may be due to inefficient breakdown of cyst wall, which consequently lead to decrease in quantity of the released DNA, in addition to disruption, and contamination of extracted DNA with impurities such as PCR inhibitors (5).
The inhibitory agents consist of fecal materials or compounds, which are remained during the process of stool preparation, cyst concentration, DNA extraction, and purification steps. Therefore, elimination of inhibitors is a must in order to obtain pure DNA. In the present study in order to overcome these complications, we included several effective approaches. Finally, the results of each DNA extraction technique were investigated by PCR amplification targeting a fragment of the conserved SSU rRNA gene with effective ability of the designed primers to specifically detect G. duodenalis. Process of cyst concentration was carried out using the sucrose flotation technique in order to decrease the fecal contaminations from purified cysts. In order to facilitate the cyst wall disruption and increase the quantity of obtained DNA, we used 7 cycles of freeze-thawing and added glass beads. Vortexing of some samples with glass beads lead to mechanically breaking the cyst wall and subsequently releasing the DNA (36).
The results of the current study have revealed the successful increase of quantity of extracted DNA and removal of inhibitors in some specimens. Using the sucrose flotation tech-nique, freeze-thawing, and adding glass beads have demonstrated the successful approaches in extraction and subsequent molecular identification of G. duodenalis (4, 22, 38). In this study, comparison of the efficiency of 3 extraction protocols on PCR fragments have shown the Phenol-Chloroform Isoamyl alcohol technique as the most effective method, especially in terms of increased quantity of the extracted DNA. This achievement was possibly due to the implementation of longer incubation time with lysis buffer containing proteinase K at 56°C, subsequently leading to more effective lysis of cysts (33). In addition, precipitation of released DNA in absolute ethanol at -20°C for 24 hours lead to increase in DNA quantity.
In order to remove polysaccharide contamination from extracted DNA cetyltrimethylammonium bromide is used and a mixture of phenol, chloroform, and Isoamyl alcohol (25:24:1) is added for isolation of proteins, lipids, carbohydrates, and cell debris from the aqueous phase containing DNA. Although CTAB and Phenol/chloroform/Isoamyl remove contaminants and PCR inhibitors, CTAB may also decrease DNA yield and incomplete removing of these compounds during washing steps can affect the quality of DNA (35). In another study, the simple phenol-chloroform method was demonstrated as the most suitable technique for DNA extraction from human samples with a sufficient quantity and quality of DNA (39).
The quality and purity of DNA are considered as the most important factors in molecular assays (34). The DNA concentration is determined using the 260 nm absorbance. Assessment of the presence of proteins in extracted DNA is performed by absorbance at 280 nm. In addition to the amount, the absorption at 230 nm can indicate that organic compounds or chaotropic salt are present. For evaluation, the purity of DNA, the A260/A280, and the A260/A230 ratios were measured and compared with standard values. The A260/280 ratio is used to assess protein contamination, with accepted value of 1.8 for pure DNA. In order to evaluate the contamination of extracted DNA with organic contaminants such as phenol and chaotropic salts, the A260/230 ratio was used, with accepted value of 2 for pure DNA (8, 34).
Our findings based on measured A260/A230 ratio as an indicator of contamination with protein or phenol and the results of PCR, demonstrated that the QIAamp DNA stool mini kit was the only successful method for DNA purification. In this commercially kit, in addition to using InhibitEX tablet for removing inhibitors, the silica membrane was commonly effective to bind and wash the extracted DNA (36). In molecular studies, less effective PCR amplification can be a consequence of the presence of the impurities in DNA template, which must be thoroughly removed during purification and washing steps.
6. Conclusions
Although in the present study the Phenol-Chloroform Isoamyl alcohol protocol was the most effective method for increasing the concentration of DNA, this technique was not completely able to remove the PCR inhibitors as described elsewhere (40). Therefore, identification and implementation of an efficient protocol for DNA extraction with the ability of obtaining sufficient quantity and quality of DNA from cysts and elimination of inhibitors are essential in molecular diagnostics studies and also in subsequent investigation of genetic diversity, taxonomy, epidemiology, and finally treatment and control strategies of G. duodenalis.