1. Background
Klebsiella pneumoniae has emerged as an important opportunistic pathogen that causes severe disease in immunocompromised patients (1-3). During the past decades, there was an increase in the incidence of infections caused by multidrug-resistant (MDR) species of Klebsiella. Infections caused by MDR K. pneumoniae are usually associated with high mortality, morbidity, healthcare costs and prolonged hospitalization. Production of extended-spectrum β-lactamase (ESBL) is the basic reason for the multidrug resistance in Gram negative bacteria throughout the world. Since 1983 when the first ESBL enzymes were described in Europe (Germany), there is an alarming increase in the incidence of ESBL-producing bacteria (4-6). ESBL-producing K. pneumoniae strains are one of the most important problems associated with nosocomial outbreak.
Among the various reported types of ESBL worldwide, CTX-M is the most prevalent and rapidly growing member of ESBL family (7, 8). CTX genes are disseminated and identified in hospitalized and outpatients worldwide. CTX enzymes are categorized into five important groups including 1, 2, 8, 9, and 25. The CTX-M-1 is the most common type of CTX-M detected in South America, Europe, and Asia (9, 10).
Genotyping methods are used to determine the clonality of the isolates in nosocomial and household outbreaks. Multilocus variable-number tandem repeat analysis (MLVA) is one of the molecular typing methods frequently used in several studies to access genetic relations and distinguishing bacterial pathogens. MLVA is a low cost, rapid, and simple method based on polymerase chain reaction (PCR) that provides the numerical data of the number of repetitions in a locus. The sequences known as variable number tandem repeat (VNTR) vary among different strains in copy number. MLVA can provide transferable information in the form of codes that can be saved in database and exchanged between different laboratories. This method, compared with other typing methods, reduced the handling of live bacteria (11-14).
2. Objectives
The current study aimed at evaluating the antimicrobial susceptibility patterns and ESBL phenotype in K. pneumoniae strains isolated from hospitalized patients. The current study also aimed at determining CTX-M-type of CTX-M-1 gene in clinical strains of K. pneumoniae isolated from Semnan, Iran to assess their genetic diversity using MLVA as a possible typing method.
3. Methods
3.1. Ethics Statement
The ethics committee of Semnan University of Medical Sciences, Semnan, Iran approved the current study protocol (IR.SEMUMS.REC 1394.84).
3.2. Sampling
The current cross sectional study was conducted at three hospitals in Semnan, Iran. A total of 110 K. pneumoniae strains were isolated from 95 urine (86.4%), 10 sputum (9.1%), and five blood (4.5%) specimens.
3.3. Bacterial Identification
In order to identify K. pneumoniae, conventional bacteriological tests including Gram stain, lactose fermentation, hydrogen gas production from glucose, FeS production, motility, indole production, sodium citrate utilization, and urea utilization were performed (15). The verified isolates were preserved at -70°C in brain-heart infusion broth with 30% (v/v) glycerol for further analysis.
3.4. Antibiotic Susceptibility Assay
The antibiotic susceptibility were determined according to the clinical and laboratory standards institute (CLSI) guidelines (16). The used antibiotic disks were cefotaxime (30 µg), nalidixic acid (30 µg), tetracycline (30 µg), ciprofloxacin (5 µg), imipenem (10 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), and amoxicillin-clavulanic acid (20/10 µg) (Rosco, Taastrup, Denmark).
3.5. Screening of ESBL Phenotype
Double-disk synergy test was recommended by CLSI (16) for phenotypic characterization of ESBL-producing isolates. This test was used to confirm ESBL production using ceftazidime (30 µg) and cefotaxime (30 µg) disks with/without clavulanate (10 µg). Klebsiella pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 were used as positive and negative controls, respectively. Positive ESBL isolates showed increase in the inhibitory zone diameter by ≥5 mm around the disks with clavulanate versus those of the ones without clavulanate.
3.6. DNA Extraction
Colonies of ESBL-producing K. pneumoniae were suspended in TE (Tris-ethylenediaminetetraacetic acid) buffer and genomic DNA was prepared by boiling method and used as the template for PCR reactions as previously described by Kuske et al. (17).
3.7. PCR Amplification to Detect CTX-M-1
The PCR was performed to detect CTX-M-1 using the following primers as previously described (18): forward primer: 5′ GACTATTCATGTTGTTGTTATTTC 3′, reverse primer: 5′ TTACAAACCGTTGGTGACG 3′, PCR product size: 923 bp. Amplification reactions were performed in a total of 25 µL volume containing 5 µL of 10X PCR buffer, 2.5 mM MgCl2, 200 mM dNTPs, 1.5 unit of Taq polymerase, 10 pM of each primer, and 1 µL of DNA template.
3.8. Multilocus Variable-Number Tandem Repeat Analysis
PCR conditions consisted of an initial activation for genotyping of isolates with positive results for CTX-M-1 gene, the eight loci, including A, D, E, H, I, J, K, and L were selected along with the primers as described by Turton (19). For this analysis, PCR was performed for each locus separately in a reaction mixture with total volume of 25 µL, containing 50 ng of DNA template, 1X PCR buffer, 2.5 mM of MgCl2, 0.2 mM of each primer, and 0.5 U of Taq DNA polymerase. The PCR condition was as follow: 95°C for 15 minutes, 35 cycles of 94°C for 30 seconds, 58°C for 90 seconds, 72°C for 90 seconds, and a final extension at 72°C for 10 minutes. After performing PCR, the size of each locus was easily determined on 1.5% gel agarose. The number of repeat for each isolate was calculated by the following formula:
Number of repeat (bp) = size of each locus (bp) - flanking regions (bp)/repeat size (bp)
3.9. Statistical Analysis
The results were documented into Microsoft excel 2007 and uploaded into the web-based MLVA plus for analyzing. Any difference in one or more VNTR locus was considered to indicate a distinct type. Simpson index of diversity for each VNTR locus was calculated using V-DICE software (Health Protection Agency, London).
4. Results
4.1. Antibiotic Susceptibility, ESBL, and CTX-M-1 Gene Detection
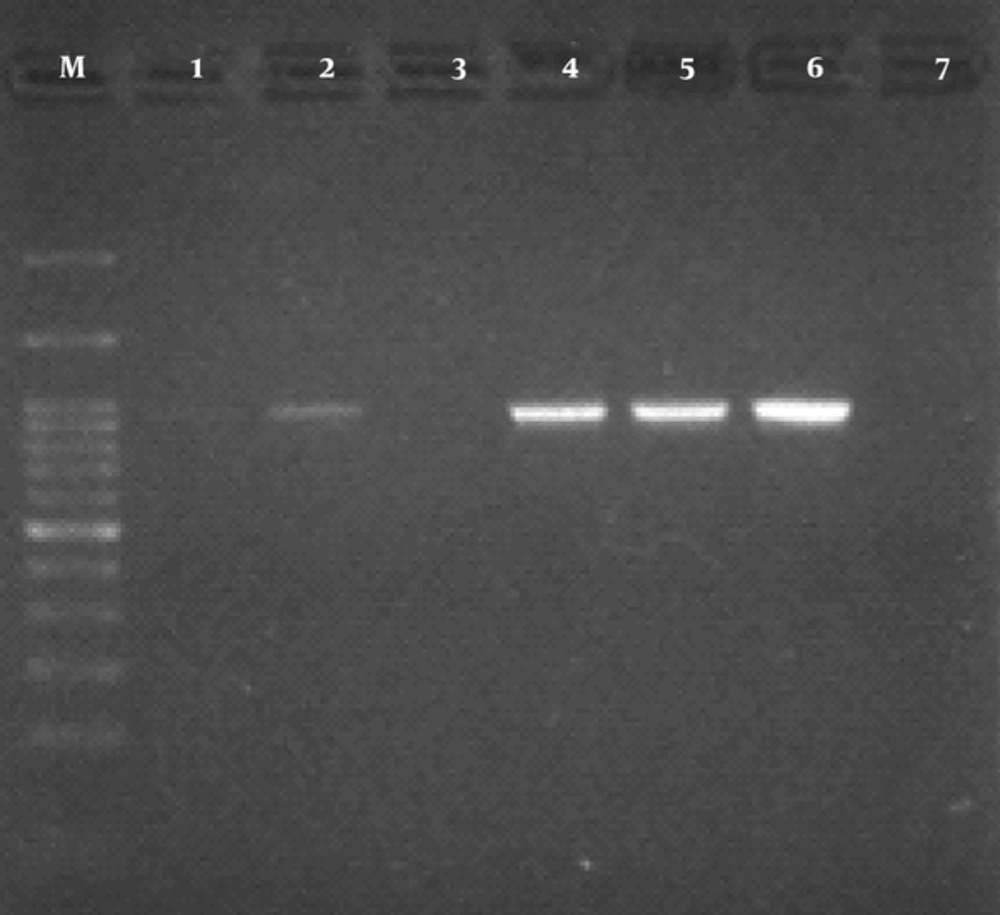
The findings of antibiotic resistant test showed that the most effective antibiotic was imipenem (84.7% susceptibility). The frequency of resistance to trimethoprim-sulfamethoxazole, cefotaxime, nalidixic acid, amoxicillin-clavulanic acid, tetracycline, and ciprofloxacin were 52.3%, 65%, 39.3%, 30.2%, 29.1%, and 27.1%, respectively (Table 1). In the current study, 70 (63.63%) isolates had ESBL positive results. The results obtained from PCR reaction using CTX-M-1 primers indicated that 42 out of 70 (60%) isolates were positive. PCR product for CTX-M-1 gene is shown in Figure 1.
| Susceptible | Intermediate | Resistant | |
|---|---|---|---|
| Amoxicillin-clavulanic acid | 67 (60.9) | 10 (9.1) | 33 (30) |
| Trimethoprim-sulfamethoxazole | 52 (47.3) | 0 | 58 (52.7) |
| Tetracycline | 77 (70) | 1 (0.9) | 32 (29.1) |
| Imipenem | 93 (84.6) | 4 (3.6) | 13 (11.8) |
| Ciprofloxacin | 79 (71.8) | 1 (0.9) | 30 (27.3) |
| Nalidixic acid | 55 (50) | 12 (10.9) | 43 (39.1) |
| Cefotaxime | 62 (56.4) | 1 (0.9) | 47 (42.7) |
4.2. Multilocus Variable-Number Tandem Repeat Analysis
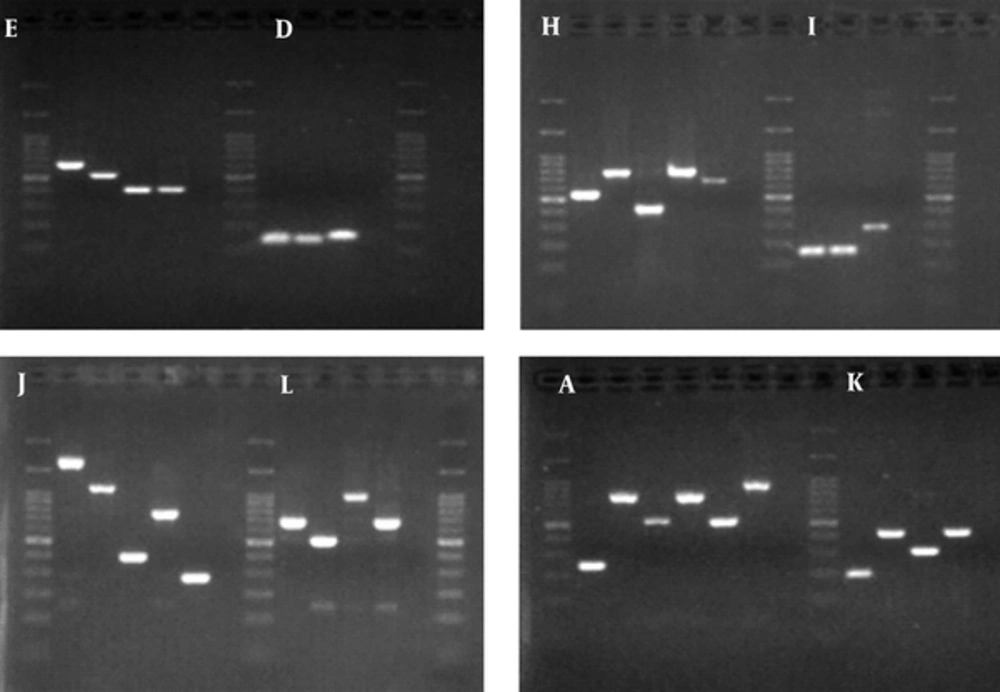
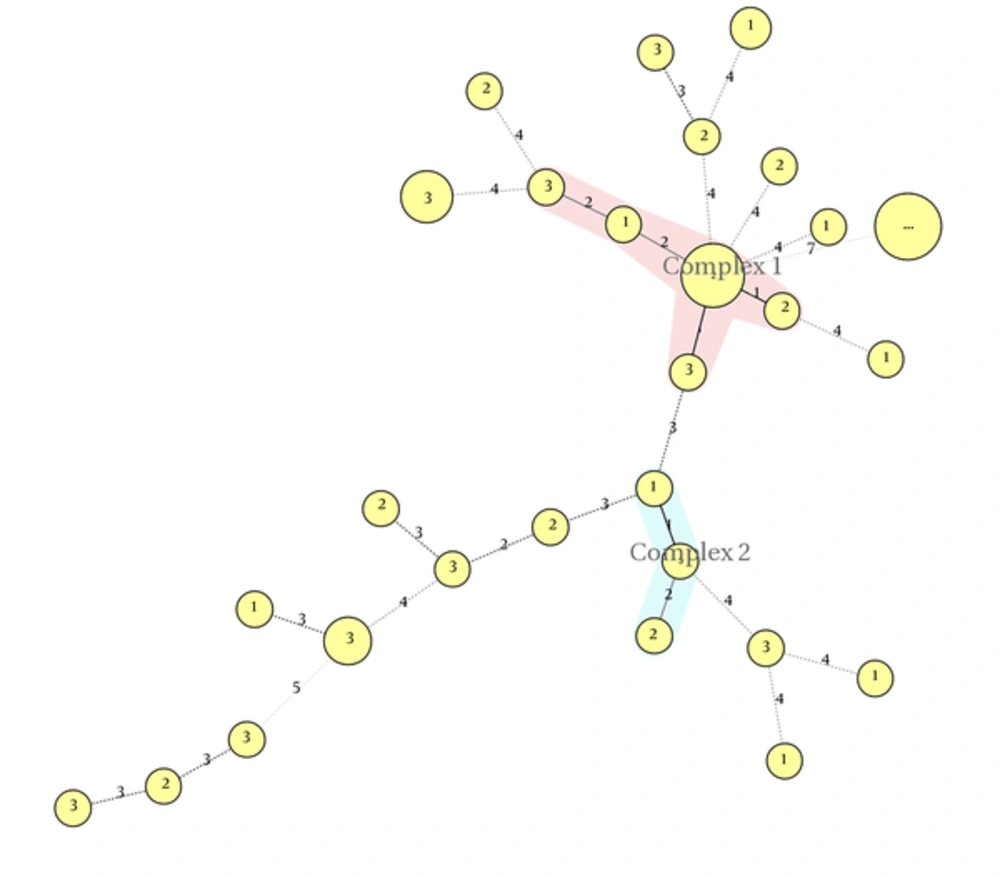
MLVA was performed on 42 K. pneumoniae isolates, using eight loci. The PCR products of these loci are shown in Figure 2. Totally, 28 MLVA genotypes were discriminated. Isolates were classified into two clonal complexes, clonal complex 1 including six isolates and clonal complex 2 including three isolate; furthermore, 19 singletons were distinguished in the isolates. Minimal spanning tree of these isolates is shown in Figure 3.
4.3. Evaluation of Clonality in the Studied Isolates
In the studied isolates, two clonal complexes (CCs) and 19 singleton isolates were categorized. The CC1 comprised six isolates and CC2 comprised only three isolates.
4.4. Reviewing Simpson Diversity Index for Each Locus
Locus J with six replicates and diversity index (DI) of 0.807 had the most differentiation power compared with the other loci used. Locus A with five replicates (DI = 0.768) ranked second in terms of differentiation power, followed by locus K with five replicates (DI = 0.750), locus H with five replicates (DI = 0.737), locus L with four replicates (DI = 0.704), and E with four replicates (DI = 0.628). Locus D with three replicates and DI = 0.475 ranked the lowest differentiation power compared with other loci.
5. Discussion
Klebsiella pneumoniae is one of the most important causative agents of several infections such as blood stream, wound, pneumonia, and urinary tract infection (20, 21). The emergence of ESBL-producing Gram-negative bacilli, especially K. pneumoniae as serious pathogen, is reported worldwide and is rapidly changing over time (22-24). In the current study, ESBL positive phenotype was observed in 42 isolates. In the current study, similar to previous studies, the most ESBL-producing K. pneumoniae were obtained from urine (25, 26). Consumption of large amount of third-generation cephalosporins by patient could describe the high prevalence of ESBL-producing K. pneumoniae (27). Since the ESBL genes are located on a mobile element, ESBL positive isolates showed a high rate of resistance to non-β-lactam antibiotics (28). Imipenem and meropenem and carbapenem were very active antibiotics against ESBL-producing K. pneumoniae. In the current study, imipenem with 84.7% susceptibility was the most effective antibiotic against K. pneumoniae.
The incidence of CTX-M-type ESBL-producing Klebsiella rapidly increased worldwide. The CTX-M enzymes are classified into five major groups based on CTX-M amino acid sequences. The occurrence of these CTX-M genotypes greatly varies among ESBL-producing isolates geographically (9, 29). Previous studies revealed that CTX-M-1 was the most common one among ESBL gene families (30-33). Forty-two out of 70 ESBL-producing Klebsiella isolates in these study carried CTX-M-1. Molecular typing is a reliable tool to study nosocomial infections. Since in recent years a significant increase of ESBL-producing K. pneumoniae is observed in hospital settings with the risk of outbreak, there is a need for a rapid and low cost typing method (34). Among the various typing methods, PFGE is accepted for typing of bacteria. However, to choose a reliable typing method in laboratory settings, several parameters such as interpretation time, costs, and complexity of performance should be considered. Since PFGE is time-consuming and technically demanding, an alternative method such as MLVA, which is easy-to-perform, cost-effective, and highly reproducible received attention (4-6, 11).
Naseer et al. applied MLVA method for genotyping E. coli carrying CTX-M-type ESBL (11). In the current study, 42 CTX-M-1 producing isolates were included to determine clonal relationship using MLVA method. In the current study, size of PCR products can be easily analyzed on agarose gel without any complicated requirement. Out of a total 42 CTX-M-1-producing K. pneumoniae isolates in the current investigation, MLVA revealed 28 distinct patterns of K. pneumoniae isolates; given that the 28 MLVA genotypes were isolated from three hospitals in one year, it can be concluded that clonal complexes with a small number of isolates in each can indicate the entry of a new bacterium in the ward. The isolates identified as singleton are possibly the bacteria that have entered the ward by the patient, the visitors or medical personnel. These isolates were not established in the ward. The current study findings demonstrated heterogeneity among CTX-M-1-producing K. pneumoniae isolates. Evaluation of diversity indexes for VNTR loci showed that VNTR-J with six different alleles was the most polymorphic and the highest diversity index of 0.807, locus D was the least variable among the current study strains with two alleles and diversity index of 0.475.
6. Conclusions
The presence of CTX-M-1 in several different MLVA types demonstrated that one specific clone was not responsible for spreading the isolates. It seems that mobility and horizontal transfer occurred among these isolates.