1. Background
Enterotoxigenic Escherichia coli (ETEC) has been reported as the most common cause of diarrhea in children below 5 years in developing countries (1). Enterotoxigenic Escherichia coli accounts for approximately 20% of all cases of diarrhea and death each year (2). As the most important cause of diarrhea in children, ETEC can be transmitted from one person to another via consumption of contaminated food or water (3).
An ETEC strain can colonize and adhere to human intestinal epithelial cells via a heterogeneous group of proteinaceous surface structures, known as colonization factors (CFs). Colonization factors can be fimbrial, nonfimbrial, or fibrillar (4). These CF structures are also referred to as CF antigen 1 (CFA/1); however, the new nomenclature suggests coli surface (CS) antigen.
So far, more than 25 CS antigens have been identified and categorized into 3 distinct families: CFA/I-like group (CFA/I, CS1, CS2, CS4, CS14, and CS17), CS5-like group (CS5, CS7, CS18, and CS20), and a distinctive group consisting of CS3, CS6, CS10, CS11, and CS12 (2, 5, 6). Following initial adhesion and colonization, ETEC strains induce diarrhea by producing heat-labile (LT) and/or heat-stable (ST) enterotoxins, which are plasmid-encoded (7). The diagnosis of ETEC infection relies on the detection of either genes or gene products in the clinical specimens (8).
Currently, LT derivatives and CFs are targets in the development of vaccines against ETEC (8). Development of these vaccines relies on the characterization of ETEC isolates in a variety of settings. Vaccine development for ETEC is dependent on an in-depth understanding of toxins and CF distribution (9). However, considering the great variability in the distribution of ETEC toxins and CFs, variations in phenotypic, genotypic, and pathogenic properties should be investigated (10). In this regard, a study carried out in South Western Nigeria reported a significant association between ST enterotoxin-producing ETEC and diarrhea (11).
2. Objectives
In view of the undocumented disease burden of ETEC-associated diarrhea in the Federal Capital Territory (FCT, Abuja, Nigeria), this study aimed to not only determine the epidemiological characteristics of ETEC strains in children with acute diarrhea, but also investigate the presence of LT and ST toxins, distribution of CFs, and antimicrobial susceptibility pattern of the strains.
3. Methods
Escherichia coli strains analyzed in this study were obtained between January and December 2012. This study included pediatric patients with diarrhea, aged 0 - 60 months, who were selected from 2 tertiary hospitals (University of Abuja Teaching Hospital, ethic code: FCT/UATH/GEN/1085/58; National Hospital of Abuja, ethic code: NHA/ADMIN/236/V.VII/805), 2 secondary hospitals (Maitama and Nyanya districts hospitals), and 1 primary health centre (Mpape Health Centre). The included facilities were all located in Abuja, Nigeria.
In total, 400 children with acute diarrhea participated in this study. The age and sex distribution of children with acute ETEC-associated diarrhea is presented in Table 1. Stool samples were collected from children ≤ 5 years, who met the inclusion criteria for diarrhea, ie, three or more loose (or watery) stools or at least 1 bloody loose stool in 24 hours. The stool specimens from diarrheic children were collected in dry, sterile, wide-mouthed, leak-proof containers. The samples were stored at 4oC in the hospitals and transferred to the microbiology laboratory in cold boxes for analysis.
The samples were evaluated for common enteric pathogens (Salmonella, Shigella, Vibrio, and Campylobacter species, rotaviruses, and ova/cyst of parasites) via conventional methods (12). All E. coli isolates were individually characterized by polymerase chain reaction (PCR), using 8 virulence gene-targeted assays to assess the presence of distinct diarrheagenic E. coli (DEC) pathotypes, as described in the literature (13). PCR-positive ETEC colonies were analyzed using ganglioside GM1 ELISA and GM1 inhibition ELISA to detect the expression of LT and ST enterotoxins, respectively, as described in the literature (14-17).
| Factors | Number of Diarrheic Subjects | Number (%) of ETEC-Positive Patients | P-Value |
|---|---|---|---|
| Age, months | 0.03 | ||
| 0 - 5 | 49 | 1 (2.0) | |
| 6 - 12 | 165 | 8 (4.8) | |
| 13 - 24 | 116 | 1 (0.9) | |
| 25 - 36 | 29 | 3 (10.3) | |
| 37 - 48 | 17 | 0 (0.0) | |
| 49 - 60 | 24 | 3 (12.5) | |
| Sex | 0.03 | ||
| Male | 180 | 10 (5.6) | |
| Female | 220 | 6 (2.7) |
aOnly 1 out of 49 subjects, aged 0 - 5 months, had ETEC infection. ETEC infection in males was significantly higher than females (P < 0.05)
3.1. DNA Extraction
A total of 109 E. coli isolates (1 isolate from each strain from every child) were grown on Luria-Bertani broth (LB broth; Sigma, St. Louis, MO, USA) overnight at 37°C. Genomic DNA was extracted according to the method described by Al-Gallas et al. in 2007 (18). Briefly, DNA was released from whole organisms by boiling. Bacteria were harvested from 1.5 mL of an overnight LB culture (Sigma, St. Louis, MO), suspended in 200 µL of sterile water, and incubated at 100oC for 15 minutes. Following lysate centrifugation, a 150-µL sample of the supernatant was stored at -20°C in the aliquot as a template DNA stock and used as the target for PCR assays.
3.2. PCR Analysis
The PCR primers for the identification of elt genes (heat-labile toxin genes) included ET-LT1 (GCGACAAATTATACCGTGCT) and ET-LT2 (CCGAATTCTGTTATATATGT) (Amersham, Piscataway, USA). The same primers were used for sta genes (heat-stable toxin genes) (18, 19). Details of primer sequences, PCR conditions, and predicted size of the amplified products for the evaluation of other virulence genes, which can distinguish DEC groups, have been described previously (Table 2) (20-23). PCR was performed in an automated thermocycler (Perkin-Elmer).
The reference strains for PCR in this study were as follows: ETEC H10407 for elt and sta genes, EHEC EDL933 (O157:H7) for stx and ehxA genes (Shiga toxin gene and enterohemolysin gene, respectively), EPEC 2348/69 (O127:H6) for eae and bfpA genes (attaching and effacing E. coli gene and bundle-forming pilus gene, respectively), EIEC 11741 for ipaH gene (invasion plasmid antigen), and EAEC 17-2 for astA and aaf-I genes (heat-stable enterotoxin I and aggregative adherence fimbriae, respectively). The nonpathogenic E. coli strain, HB101, was used as the negative control.
| Gene | Primer | Oligonucleotide Sequence (5’-3’) | PCR product, bpa | PCR Condition | Reference |
|---|---|---|---|---|---|
| Elt | 708 | 94°C, 1 min; 56°C, 2 min; 72°C, 1 min | (19) | ||
| ET-LT1 | GCGACAAATTATACCGTGCT | ||||
| ET-LT2 | CCGAATTCTGTTATATATGT | ||||
| Sta | 180 | 94°C, 30 sec; 55°C, 90 sec; 72°C, 90 sec | (20) | ||
| ST5’ | ATTTTTCTTTCTGTATTGTCTT | ||||
| ST3’ | CACCCGGTACAAGCAGGATT | ||||
| Eae | 790 | 94°C, 1 min; 55°C, 1 min; 72°C, 1 min | (21) | ||
| fM1 | CATTATGGAACGGCAGAGGT | ||||
| rYu4 | ATCTTCTGCGTACTGCGTTCA | ||||
| bfpA | 324 | 94°C, 30 sec; 43°C, 30 sec; 72°C, 1 min | (19) | ||
| EP1 | CAATGGTGCTTGCGCTTGCT | ||||
| EP2 | GCCGCTTTATCCAACCTGGT | ||||
| Stx | 900 | 94°C, 1 min; 43°C, 90 sec; 72°C, 90 sec | (22) | ||
| LIN5’ | GAACGAAATAATTTATATGT | ||||
| LIN3’ | TTTGATTGTTACAGTCAT | ||||
| ehxA | 321 | 94°C, 90 sec; 55°C, 90 sec; 72°C, 2 min | (20) | ||
| RH35 | CACACGGAGCTTATAATATTCTGTCA | ||||
| RH37 | AATGTTATCCCATTGACATCATTTGACT | ||||
| IpaH | 424 | 94°C, 1 min; 56°C, 2 min; 72°C 1 min | (19) | ||
| EI-1 | GCTGGAAAAACTCAGTGCCT | ||||
| EI-2 | CAGTCCGTAAATTCATTCT | ||||
| astA | 111 | 95°C, 30 sec; 55°C, 30 sec; 72°C, 30 sec | (23) | ||
| Eg1 | CCATCAACACAGTATATCCGA | ||||
| Eg2 | GGTCGCGAGTGACGGCTTTGT | ||||
| aaf/I | 432 | 95°C, 30 sec; 55°C, 30 sec; 72°C, 30 sec | (23) | ||
| AAF/I-1 | GCGTTAGAAAGACCTCCAATA | ||||
| AAF/I-2 | GCCGGATCCTTAAAAATTAATTCCGGC |
aThe PCR cycles consisted of denaturation, annealing, and extension
3.3. GM1 ELISA and CF Immunodot Blot Assay
Strains identified as ETEC by PCR were confirmed by ganglioside GM1 ELISA. GM1 inhibition ELISA was performed to detect the expression of LT and ST enterotoxins, respectively as described in the literature (14, 15). Subsequently, two colonies from each strain, which were confirmed as ETEC via ELISA, were subcultured on CF agar with and without bile salts (Difco, Detroit, MI) and incubated at 37°C overnight, as described in the literature (17, 24). The expression of phenotypic CFs was tested by colony immunodot blot assay, using monoclonal antibodies against CFA/I, CS1–CS7, CFA/III (CS8), CS12 (PCFO159), CS14 (PCFO166), and CS17 (25-27).
3.4. Determination of Hemagglutination (HA) Activity
The presence of HA was tested for all GM1 ELISA-confirmed ETEC strains, using the methods described by Evans et al. (28). Briefly, a colony from each isolate of CFA culture was suspended in 10 µL of saline on a glass slide in a repetitive order. Each slide was added 10 µL of 3% (v/v) human, bovine, chicken, and guinea pig erythrocyte suspension in saline, with or without 1% D-mannose. The slide was rotated back and forth for mixing for 5 minutes. The presence or absence of HA was scored as (-) or (+). HA was considered mannose-sensitive (MSHA) if only detected in the erythrocyte suspension without 1% D-mannose. On the other hand, HA was considered mannose–resistant (MRHA) if detected in the presence or absence of 1% D-mannose.
3.5. Serotyping of E. coli
The serological characterization of 16 ETEC strains into different somatic (O), flagella (H), and K1 antigens was performed at the Identification and Serotyping Laboratory of the Enteric Diseases Program, National Microbiology Laboratory, Canadian Science Centre for Human and Animal Health Public Health Agency of Canada (1015 Arlington St., Winnipeg, Manitoba R3E 3R2, Canada). The procedures were performed as described by Orskov and Orskov (1984) (29-31).
3.6. Antimicrobial Susceptibility
The characterized ETEC strains (n, 16) were tested for susceptibility to the following antibiotics: nalidixic acid (30 µg), ciprofloxacin (5 µg), cephalexin (30 µg), cefuroxime (30 µg), amoxicillin (25 µg), ceftriaxone (30 µg), and amoxicillin-clavulanic acid (30 µg) (Oxoid, Basingstoke, UK), using the standard Kirby-Bauer disc diffusion susceptibility method. In addition, Escherichia coli ATCC 25922 was used as the control.
Multidrug resistance (MDR) was defined as resistance to ≥ 2 classes of antimicrobial agents (32). The minimum inhibitory concentrations (MICs) of the isolates were determined by the broth microdilution method (Sensititre Automated Microbiology System; Trek Diagnostic Systems Ltd., Westlake, OH, USA). Using the Clinical & Laboratory Standards Institute (CLSI) guidelines, each organism was classified as either resistant or susceptible (33).
3.7. Ethics statement
Informed consent and ethical approval were obtained from the subjects and institutional ethics committees of the University of Abuja Teaching Hospital (Gwagwalada, Abuja) and National Hospital of Abuja, respectively.
3.8. Statistical Analysis
Statistical analysis was performed using SPSS version 16 with Fisher’s exact test. P-value below 0.05 was considered statistically significant.
4. Results
In this study, we evaluated the prevalence of ETEC, its toxins, CFAs, and O:H serotypes associated with childhood diarrhea in FCT-Abuja, Nigeria. Enterotoxigenic Escherichia coli associated diarrhea was observed in all age groups, except children aged 37 - 48 months, from whom no ETEC strain was isolated. The association between ETEC and age group of 49 - 60 months was significant (P < 0.05). The prevalence of ETEC infection was higher in male subjects (5.6%; 10/180), compared to females (2.7%; 6/220) (P < 0.05; Table 1).
4.1. Detection of Toxins and CFs
The phenotypic characteristics of ETEC strains are illustrated in Table 3. The expression of CFA/CS antigen in ETEC strains varied significantly. In total, 16 ETEC strains expressed CFA; four (25.0%) strains expressed CS2, 2 (12.5%) strains expressed CS3, and 1 (6.3%) strain expressed CFA1. CFA/CS was not expressed in 9 (56.3% ETEC strains (5 LT and 4 ST strains). The association between CFA/CS and ETEC strains, positive for both LT and ST, was significant (P < 0.05). The frequencies of ETEC-ST, ETEC-LT, and ST-LT strains were 6/16 (37.5%), 6/16 (37.5%), and 4/16 (25%), respectively. Overall, 43.7% of ETEC isolates expressed CFs, while ETEC strains with no detectable CFA/CS consisted of LT (31.3%) and ST (25.0%) strains.
| Isolate ID No. | Enterotoxin Type | CFA/CS | HEp-2 cell Adherence Pattern | Serotype |
|---|---|---|---|---|
| 006 | st | - | NA | Escherichia coli O117:HNM |
| 013 | st | CS2 | DA | E. coli O86:H18 |
| 014 | lt | CS3 | DA | E. coli O8:H9 |
| 015 | st | - | NA | E. coli ORough:HNM |
| 034 | lt | - | NA | E. coli OUT:HNM |
| 140 | st | CFA/I | DA | E. coli O15:H11 |
| 189 | st/lt | CS2/CS3 | DA | E. coli O8:H9 |
| 190 | st/lt | CS3/CS2 | DA | E. coli O8:H9 |
| 234 | st | - | NA | E. coli O86:H30 |
| 283 | st | - | NA | E. coli ORough:HNM |
| 290 | lt | - | NA | E. coli O9:HUT |
| 297 | lt | - | NA | E. coli O9:HUT |
| 344 | st/lt | CS2/CS3 | DA | E. coli O8:H9 |
| 345 | st/lt | CS2/CS3 | DA | E. coli O8:H9 |
| 354 | lt | - | NA | E. coli OUT:HNM |
| 361 | lt | - | NA | E. coli OUT:HNM |
Abbreviations: DA, diffuse adherence; NA, nonadherence; CFA/I, colonization factor antigen I; CS2, coli surface 2; CS3, coli surface 3; HNM, flagellar and nonmotile; HUT, flagellar and untypeable; OUT, O-untypeable
4.2. Demographic and Seasonal Distribution of ETEC
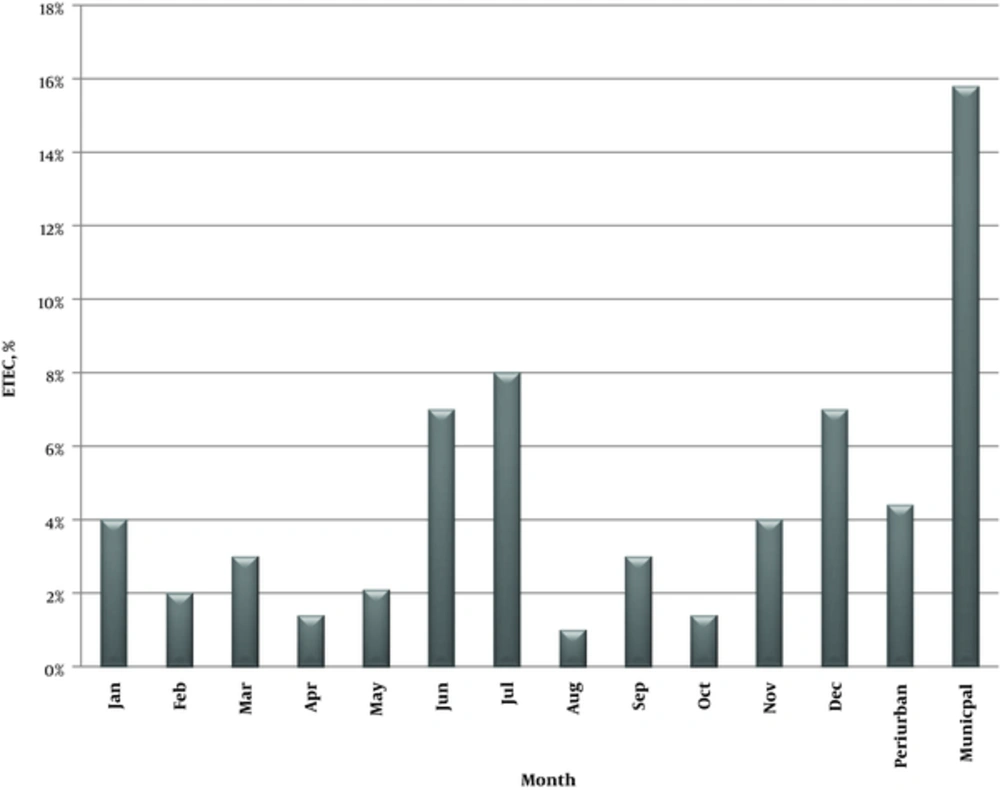
Demographic distribution and seasonal variation of ETEC isolates from children with acute diarrhea in Abuja, Nigeria are presented in Figure 1. Enterotoxigenic Escherichia coli associated diarrhea was reported throughout the study (January-December). The seasonal patterns in this study indicated that ETEC-associated diarrheal infections peaked in July (7.2%), although the difference was not significant (P > 0.05). The second highest prevalence of infection was recorded in December (6.6%), while lower prevalence rates were observed in August and October.
4.3. Determination of Antimicrobial Resistance Patterns of ETEC Strains
The antimicrobial resistance patterns of ETEC strains, obtained by disc diffusion susceptibility method, are illustrated in Table 4. All 16 ETEC strains were resistant to at least 1 antimicrobial agent, whereas none exhibited resistance to the tested antimicrobials according to the current CLSI breakpoints. There were 7 different antimicrobial resistance patterns among the ETEC strains. Overall, MDR was observed in 5/16 (31.3%) ETEC strains.
| ETEC Toxin Type | Total Number of Strains | Antibiotic Resistance Profile | Frequency | |
|---|---|---|---|---|
| No. | Percentage | |||
| St | 6 | AMX | 1 | 16.7 |
| AMX, AUG | 1 | 16.7 | ||
| AUG, CEPH | 1 | 16.7 | ||
| NAL, AMX, AUG, CEPH, CEFU | 1 | 16.7 | ||
| NAL, CIP, AMX, AUG, CEPH, CEFU | 2 | 33.3 | ||
| Lt | 6 | AMX, AUG | 1 | 16.7 |
| AMX, AUG, CEPH | 3 | 50.0 | ||
| AMX, AUG, CEPH, CEFU | 2 | 33.3 | ||
| st/lt | 4 | AMX, AUG, CEPH | 3 | 75.0 |
| NAL, CIP, AMX, AUG, CEPH, CEFU | 1 | 25.0 | ||
The antibiotic resistance profiles showed that 2 (33.3%) out of 3 ETEC strains with resistance patterns of NAL, nalidixic acid; CIP, ciprofloxacin; AMX, amoxicillin; AUG, augmentin; CEPH, cephalexin; CEFU, cefuroxime to 6 out of 7 tested antibiotics possessed ST toxin type, whereas the remainder possessed ST/LT toxin
5. Discussion
Enterotoxigenic Escherichia coli has been identified as a major etiological agent of diarrheal disease among travelers and children below 5 years in developing countries. The prevalence of ETEC was reported to be 10.3% and 12.5% in children aged 25 - 36 and 49 - 60 months, respectively. These rates are higher than previous reports from South Western Nigeria (2.4%) (11). Similarly, recent studies from other developing countries reported an ETEC prevalence of 5.2% in children below 2 years in Peru (8) and 9.2% in Brazil (34).
In the present study, the prevalence of ETEC in children aged 49 - 60 months (12.5%) is not in agreement with the general belief that the prevalence of ETEC infection in developing countries decreases after 5 years of age (2). However, this finding is consistent with a recent report, indicating a significantly higher ETEC isolation rate among older infants (8). The discrepancy between our findings and other reports may be attributable to variations in geographical factors and target populations.
The sta and elt genes were equally distributed among ETEC isolates (37.5%); this finding is not in agreement with previous studies, which reported ST dominance in Nigeria (11) and Jakarta, Indonesia (35). However, the high prevalence of LT- and ST-producing ETEC isolates in this study was similar to the distribution of enterotoxins in other studies on diarrhea among children, including those performed in Egypt, Tunis, and Bolivia (10, 18, 36).
Studies from different parts of the world have reported the phenotypic diversity of ETEC with regard to its potential for the combination of serotypes, toxins, and CFs (1, 8, 18). The prevalence of CF ETEC strains in this study (43.7%) is lower than recent reports of ETEC strains isolated from diarrheal specimens in Egypt (64%), Peru (65%), and Bolivia (55%) (1, 8, 31). The differences in CF detection in these studies may be due to variations in the methodologies of dot blot assay and multiplex PCR for detection.
Detection of no CFA/CS in 56.3% of ETEC strains in this study is consistent with previous research, which indicated the inability to detect known CFs in approximately 30 - 50% of ETEC strains (37). This finding has been attributed to the absence of CFs, their loss, or lack of specific tools for detection (37). Moreover, lack of a defined CF has been reported to be mainly related to LT strains (10, 33), which is in agreement with the present results.
Colonization factors are usually more prevalent in ST-positive strains (26). However, we detected CFs more in ST- and LT-positive ETEC strains, which is consistent with reports from peri-urban regions of Egypt (3). Since human infection by CF-negative ETEC strains continues to be reported, further studies are needed to determine the virulence and involvement of CF-negative ETEC strains in diarrheal disease.
In this study, a specific toxin-serotype association was observed, particularly with serotype O8: H9 among the ETEC strains. This serotype has been reported to be associated with LT-positive strains (36, 38). However, in the present study, this serotype was observed in 5 strains, all isolated from different patients; four were associated with ST/LT-positive strains, and only 1 was LT-toxin positive.
The HA assay of E. coli isolates revealed different patterns with various erythrocytes. Based on the HA study, 7 (43.8%) ETEC strains were MRHA, while only 1 (6.3%) was MSHA. Previous studies in Argentina and Kuala-Lumpur reported the absence of HA in 46.2% and 62.0% of ETEC strains, respectively (39, 40). In the present study, 9 (56.3%) ETEC strains failed to demonstrate any type of HA, which is in agreement with previous reports. The absence of HA in a significant proportion of ETEC isolates can be attributed to the loss of CFA encoding genes or the presence of other adhesions not associated with HA (32); the foregoing may be the case with ETEC strains in this study.
CFA I and CFA II (CS2 and CS3), which constitute only 18% of oral inactivated vaccine candidates and have been evaluated by experts, are the actual CFAs in FCT-Abuja, Nigeria. In Bangladesh, the prevalence of toxins and CFs in bacterial components of candidate ETEC vaccines has been estimated at 75% (2). This implies that the formalin-inactivated ETEC vaccine may be of limited use, as CFA I and CFA II (CS2 and CS3) are found in the bacterial components of current candidate ETEC vaccines in FCT-Abuja, Nigeria. Therefore, this vaccine may not confer protection against a number of ETEC strains prevalent in this region.
Since ETEC toxin and CF variability is indispensable (especially as formulation of ETEC vaccines is mainly based on LT toxins and common CFs) (41), it is pertinent to use a combination of maximally antigenic vaccine preparations and regimens for delivery. This can produce an optimal immune response to ETEC strains, infecting infants and children in developing countries (6). It also ensures that ETEC strains are not refractory to these vaccine preparations and underscores the relevance of our study as the first analysis and description of CFs among ETEC strains in Nigeria.
Seasonal variations in this study indicated that ETEC disease peaked during rainy seasons, although the prevalence was not significantly higher than dry seasons. Although this finding is consistent with the usual peak of infections during transition to warm and rainy seasons in tropical regions, it disagrees with the unusual seasonality for ETEC strains recently reported in Bolivia (1).
Antimicrobial resistance by ETEC strains indicated that the amoxicillin-augmentin-cephalexin resistance profile was the most common resistance pattern, while ETEC strains were susceptible to ceftriaxone. We also detected resistance to nalidixic acid and ciprofloxacin. Overall, E. coli resistance to quinolone has been mainly associated with mutations in gyrA and parC genes. Strains resistant to nalidixic acid are likely to have at least 1 mutation of gyrA gene. Although MDR was 31.3% among ETEC strains, no extended-spectrum beta-lactamase (ESBL producers were detected among these strains. This finding is similar to a study by Al-Gallas et al. in 2007 (18). However, it should be noted that in children with acute gastroenteritis, use of specific antimicrobial drugs should be limited to well-defined bacterial agents (8).
6. Conclusion
The present analysis of ETEC-associated diarrhea among children in FCT-Abuja, Nigeria confirms that ETEC is still a major cause of diarrhea in developing countries. The findings accentuate the need for sustained nationwide epidemiological ETEC studies, as there may be a loss of CF plasmids during storage, subculturing, and processing. It should be noted that some adhesions with the potential to extend the protective spectra of future candidate vaccines might have been missed from the assays in Nigeria. This study highlights the need for major efforts towards the design of more effective vaccines for the prevention of ETEC infections, especially in endemic areas. Overall, the present findings may be useful in this area. This study also established the role of ETEC in childhood diarrheal infections in FCT-Abuja, Nigeria. The findings underscore the need for continuous identification of more commonly expressed ETEC bacterial components as a framework for evaluating potential ETEC vaccines in Nigeria.