1. Background
Urinary tract infection (UTI) is known as an infection that affects 150 million people annually and the second most common infection reported in Iran and the world. It causes seven million visits by doctors due to UTI each year (1, 2). Klebsiella pneumoniae and Escherichia coli among the Enterobacteriaceae family are more likely to be acquired infections and hospital infections than the rest of the family (3). Klebsiella is an opportunistic organism that causes hospital infection and acquired community infection, such as septicemia, pneumonia, and urinary tract infections, soft tissue infections, liver damage, abscess, and meningitis (4). The importance of this bacterium as a human pathogen is the development of hospital infections and the involvement of the patients with immunodeficiency or underlying diseases such as diabetes mellitus and chronic pulmonary disorders (5).
The emergence of antibiotics such as cephalosporin and aztreonam and their increased consumption have led to the emergence of a new group of β-lactamase enzymes called broad-spectrum β-lactamases (ESBLs) (6). Although carbapenems are preferred choice for infections nowadays, unfortunately, many Gram-negative bacteria with multi-drug resistance (MDR) are also resistant to carbapenems (7). The β-lactamase genes are transmitted vertically and horizontally between bacteria. There are more than 300 sub-groups of ESBLs, which mutations in their genes led to the production of new β-lactamases capable of hydrolyzing third-generation cephalosporins and aztreonam (8). So far, 340 β-lactamase enzymes have been identified and classified according to the classification of the ambler to four categories A, B, C, and D, which A, C, and D have the active site for serine, while class B requires zinc ion for its activity, and class D includes a group that is naturally similar to class A and C enzymes, which are the same as oxacillinase (9).
OXA beta-lactamases are found mainly in Enterobacteriaceae and Pseudomonas aeruginosa and belong to the molecular D class. They are responsible for resistance to ampicillin and cefalotin and are characterized by high hydrolytic activity against oxacillin and flucloxacillin, weakly inhibited by clavulanic acid and are often resistant to ceftazidime and low resistant to carbapenems (10, 11). For the first time, PER gene in a clinical specimen of a patient in a French hospital has been reported from P. aeruginosa. PER β-lactamases have high resistance to ceftazidime, cefotaxime, and monobactam; additionally, they can cause low resistance to carbapenems and cephamycin. They are well controlled by clavulanic acid and tazobactam (12). Therefore, rapid detection and identification of these resistant strains help to adopt an appropriate and concise treatment and helps to control infection and prevent the further development of them (13-15) It is, therefore, necessary to determine the pattern of antibiotic resistance of bacteria in each region. Considering the prevalence of ESBLs in Klebsiella in every region makes possible for the clinicians to choose effective antibiotics in order to quickly eradicate infections caused by these bacteria.
2. Objectives
The aim of this study was to determine β-lactamase enzymes, minimum inhibitory concentration (MIC) for ceftazidime, cefotaxime, and ceftriaxone and determination of the frequency of β-lactamase OXA-10 and PER genes in Klebsiella strains isolated from UTIs.
3. Methods
3.1. Bacterial Isolates
In this descriptive-analytical study, 90 isolates of Klebsiella were collected from patients with UTI in two inpatient (n = 45) and outpatient (n = 45) groups in 2016. Inpatient samples included those taken from patients who had been diagnosed with UTI 48 - 72 hours after being admitted to the hospital. Outpatient samples were also prepared from the patients who had not a hospital stay in the past month. Klebsiella isolates were recognized through microscopy, Gram stain, culture, and biochemical tests. Eosin methylene blue media, MacConkey agar, and Blood agar (Merck Co., Germany) were used for culture. Uropathogenic microorganisms were known via phenotypic tests of exact culture and typical phenotypic structures. Bacterial isolates were then recognized by Gram staining and a series of typical biochemical tests.
3.2. Antimicrobial Susceptibility Testing
An antibiotic susceptibility test was done by the disk diffusion method (Kirby Bauer), according to the clinical laboratory standards institute criteria (CLSI) on Mueller-Hinton agar (Merck, Germany). The discs of antibacterial used in this study contained amikacin (30 ug), ceftazidime (30 ug), ciprofloxacin (5 ug), cefazolin (30 ug), imipenem (10 ug), co-trimoxazole (25 ug), and nalidixic acid (30 ug) by disc diffusion method.
3.3. Minimum Inhibitory Concentration and Phenotypic Confirmatory Tests
To determine the production of broad-spectrum β-lactamase enzymes, phenotypic confirmatory tests were performed, including ceftazidime (30 ug) and ceftazidime/clavulanic acid (10 ug) disc. If the diameter of halo ceftazidime/clavulanic acid was greater than 5mm in comparison to ceftazidime, ESBL production was confirmed. Then MIC for ceftazidime, cefotaxime, and ceftriaxone was determined by E-test tape method. The E-test has been reported to be a simple and correct different method for determining the antimicrobial susceptibility of different microorganisms. The MIC for ceftazidime, cefotaxime, and ceftriaxone was determined according to the general committee’s clinical laboratory standard criteria for phenotypic ESBL-producing Klebsiella isolates using the E-test strip (16, 17).
3.4. DNA Extraction and PCR
The plasmid DNA was extracted by using a fermentas plasmid kit. The PCR test was performed using specific primers for OXA-10 and PER genes (18). The 16srRNA gene was also used for internal control. Positive PCR control is Klebsiella pneumoniae ATCC 700603 (19). The sequence of the primers was shown in Table 1.
| Primers | Sequences | Genes | Size (bp) |
|---|---|---|---|
| OXA-10 | blaOXA-10 | 720 | |
| F | GTCTTTCGAGTACGGCATTA | ||
| R | ATTTTCTTAGCGGCAACTTAC | ||
| PER | blaPER | 927 | |
| F | ATGAATGTCATCACAAAATG | ||
| R | TCAATCCGGACTCACT | ||
| 16SrRNA | Internal | 420 | |
| F | AGGCCTTCGGGTTGTAAAGT | ||
| R | ACCTCCAAGTCGACATCGTT |
4. Results
In this study, from 45 outpatients, 18 were female and 27 were male. Of the 45 inpatients, 22 were female and 23 were male. In terms of age, the minimum age was 6 years and the maximum age was 64 years. From 90 isolates of Klebsiella, 82 isolates belonged to K. pneumoniae species and 8 isolates to K. oxytokka species. In this study, out of 45 inpatient samples, 40 isolates of K. pneumoniae and 5 isolates of K. oxytokka were isolated. Also, 42 isolates of K. pneumoniae and 3 isolates of K. oxytokka were gathered from outpatient infections. The susceptibility pattern of different antibiotics, as well as the percentage of resistance in inpatients and outpatients, were presented in Table 2. As shown in Table 2, the highest antibiotic resistance in inpatient and outpatients was related to antibiotic sulfamethoxazole-trimethoprim, which was 86.6% and 66.7% in the inpatient and outpatient, respectively.
| Antibiotics | Inpatient | Outpatient | P Value | ||||
|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | ||
| Amikacin | 34 (75.6) | 8 (17.7) | 3 (6.7) | 41 (91.1) | 3 (6.7) | 1 (2.2) | < 0.001 |
| Ceftazidime | 12 (26.6) | 3 (6.7) | 30 (66.6) | 32 (71.1) | 5 (11.1) | 8 (17.8) | < 0.001 |
| Ciprofloxacin | 20 (44.4) | 10 (22.2) | 15 (34.4) | 32 (71.1) | 4 (8.9) | 9 (20) | < 0.001 |
| Imipenem | 18 (40) | 3 (6.7) | 24 (53.3) | 30 (66.7) | 5 (11.1) | 10 (22.2) | < 0.001 |
| Ceftriaxone | 11 (24.4) | 2 (4.5) | 32 (71.1) | 31 (68.8) | 2 (4.5) | 12 (26.7) | < 0.001 |
| Cefotaxime | 13 (29) | 1 (2.2) | 31 (68.8) | 30 (66.7) | 6 (13.3) | 9 (20) | < 0.001 |
| Sulfamethoxazole-trimethoprim | 4 (9) | 2 (4.4) | 39 (86.6) | 10 (22.2) | 5 (11.1) | 30 (66.7) | < 0.001 |
The results indicated that antibiotic resistance was higher in inpatients compared to outpatients. Also, based on the results, the lowest antibiotic resistance was found in amikacin antibiotics in inpatients and outpatients, which was 6.7% and 2.2%, respectively. Antibiotic resistance patterns based on MIC were provided in Tables 3 - 5. Based on Fischer’s exact test, P value < 0.001 was considered significant. The results showed that there was a significant relationship between increasing the MIC of ceftazidime, cefotaxime, and ceftriaxone and Klebsiella strains isolated from UTI. A confirmatory phenotypic test was used to confirm the production of broad-spectrum β-lactamase enzymes.
| Group | Ceftriaxone | Totals | P Value | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Inpatient | 11 | 2 | 32 | 45 | < 0.001 |
| Outpatient | 31 | 2 | 12 | 45 | |
| Totals | 42 | 4 | 44 | 90 | |
| Group | Cefotaxime | Totals | P Value | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Inpatient | 13 | 1 | 31 | 45 | < 0.001 |
| Outpatient | 30 | 6 | 9 | 45 | |
| Totals | 43 | 7 | 40 | 90 | |
| Group | Ceftazidime | Totals | P Value | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Inpatient | 12 | 3 | 30 | 45 | < 0.001 |
| Outpatient | 32 | 5 | 8 | 45 | |
| Totals | 44 | 8 | 38 | 90 | |
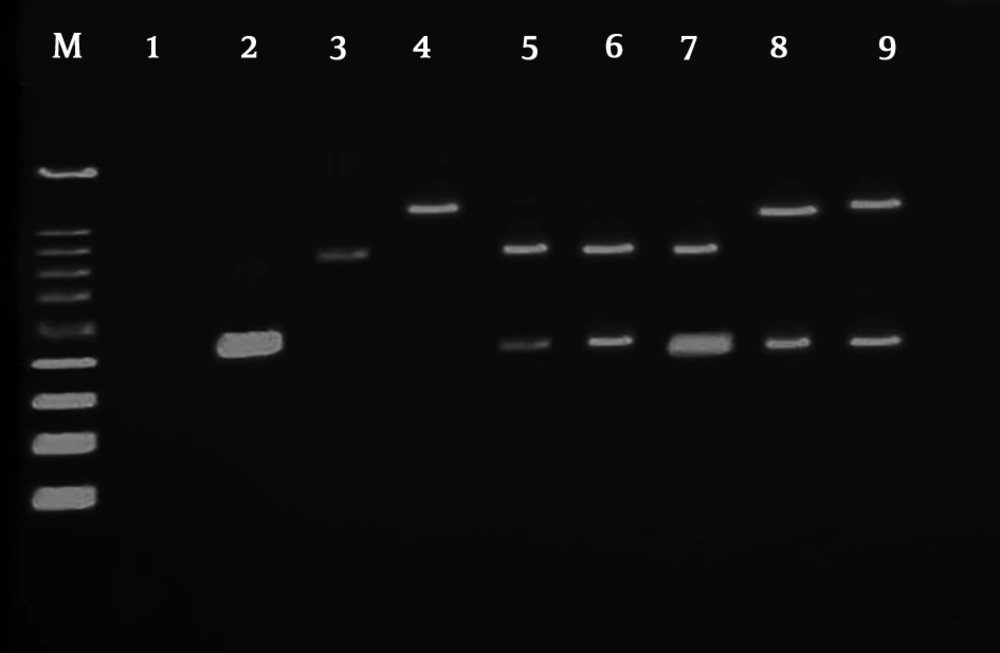
Based on the results of this test, 30 isolates of inpatient samples and 8 isolates of outpatients were positive for producing of ESBLs. PCR method was used to determine the frequency of β-lactamase genes. After the completion of the PCR steps, electrophoresis was applied to agarose gel to observe and separate the bands. After completion of electrophoresis, the agarose gel was stained (Figure 1). Accordingly, the frequency of β-lactamase OXA-10 and PER genes in Tables 6 and 7 is shown in two groups of inpatient and outpatient specimens. According to the results, there was a significant relationship between the presence of β-lactamase genes in inpatients and outpatients.
| Group | Positive | Negativ | P Value |
|---|---|---|---|
| OXA-10 | < 0.001 | ||
| Inpatient | 30 | 15 | |
| Outpatient | 4 | 51 | |
| Totals | 34 | 44 | |
| PER | < 0.001 | ||
| Inpatient | 10 | 35 | |
| Outpatient | 1 | 44 | |
| Totals | 11 | 79 |
| Group | ESBL | Totals | P Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Inpatient | 30 | 15 | 45 | < 0.001 |
| Outpatient | 8 | 37 | 45 | |
| Totals | 38 | 52 | 90 | |
5. Discussion
Klebsiella pneumoniae is one of the most important bacteria that we are witness to the emergence and development of resistant strains with MDR in recent decades due to excessive and incorrect consumption of antibiotics. Therefore, in such a situation, rapid diagnosis of MDR strains and the provision of appropriate therapeutic approach for both the patient and the treatment system is very important. The release of drug resistance factors among Gram-negative bacteria increases the resistance of these microorganisms to antimicrobial antibiotics, the persistence of these infections, and the prolongation of the patient’s therapeutic procedures (20-22).
The present study was carried out to determine the antibiotic susceptibility pattern on Klebsiella isolates from urinary tract infections. The highest antibiotic resistance is in the patients receiving trimethoprim-sulfamethoxazole antibiotics. The results indicated that antibiotic resistance was high in inpatients compared to outpatients. Also, based on the results, the lowest antibiotic resistance was found in amikacin antibiotic in the inpatients and outpatients. Current findings indicated that antibiotic resistance is higher in inpatients than outpatients. Saeidi et al., in 2014, conducted an antibiotic resistance study for ceftriaxone (100%) and ceftazidime (100%), which was higher than the resistance pattern in this study for these antibiotics (20). In a study conducted by Eftekhar et al., in 2012, antibiotic resistance was evaluated to third-generation cephalosporins such as ceftazidime, ceftriaxone plus other antibiotics such as amikacin and nitrofurantoin, with 41%, 2%, 37%, 41%, and 49%. The percentage was reported to be similar to the antibiotic resistance pattern in this study (23).
In a study conducted by Gholipour et al. in Isfahan, the results of the antibiotic resistance pattern for nalidixic acid, gentamicin, cefepime, and amikacin were lower than those reported in this study, which is consistent with our study for the low resistance to amikacin (24). The confirmed phenotypic test results in this study indicated that 66.6% and 17.7% of the isolates were able to produce ESBL among inpatients and outpatients. According to a study conducted by Jalalpoor and Mobasherizadeh, in Esfahan in 2010, the prevalence of ESBLs in inpatients was 64% with a high incidence of ESBLs in this study for inpatients (25).
Generally, the prevalence of ESBL production is different in each region and country due to medical staff’s performance and antibiotic use patterns, infection control measured in the hospital, and epidemiologic factors. It seems that the selective pressure exerted by the use of broad-spectrum antibiotics and the misuse of antibiotics with the high potential for release of plasmid genes of group A β-Lactamase (by conjugation) are main factors in the development of ESBL-producing strains in this region over time (26, 27). Feizabadi et al. reported the prevalence of these enzymes as 44.5% and 72.1% in two separate studies in Tehran (28, 29). In South Korea, the prevalence of ESBLs was 30%, in India, 68%, in Taiwan, 28.4%, and in the United States, 44% (30-33).In the present study, the frequency of β-lactamase OXA-10 and PER genes were 66.6% and 8.8%, respectively, in inpatients and 22.2% and 2.2%, respectively, in outpatients. In previous studies, the frequency of the OXA gene in Klebsiella and other bacteria from the Enterobacteriaceae family was more than PER and their frequency was higher in inpatients than outpatients. In a study conducted by Jabalameli et al. in 2011, and 112 samples were isolated, the prevalence of β-lactamase genes, including OXA-10, PER, and VEB was reported as 70%, 50%, and 31%, respectively. This research is consistent with our study for the prevalence of OXA-10 gene, which is more than PER (34). As in the study of Farshadzadeh et al. in 2014, the prevalence of PER, OXA, and CTX-M genes in 176 isolated strains from inpatients was 55%, 66% and 1%, respectively (10). In a study by Luo et al. in 2011, to investigate the prevalence of broad-spectrum β-lactamases in 59 isolates of Klebsiella, the frequency of OXA, SHV, and CTX-M genes was reported to be 22.1%, 91.5%, and 59.9%, respectively. The prevalence of OXA-10 gene is lower than that of the present study, which can be explained by the fact that Luo et al. worked on fewer isolates (35).
The prevalence of PER in bacteria such as Klebsiella and E. coli is usually as low as in the following studies. However, the prevalence is much higher in other bacteria such as Pseudomonas and Acinetobacter. In this regard, in a study conducted by Shacheraghi et al. in 2010 in Iran, 120 samples of P. aeruginosa were isolated from burn infections, which the prevalence of β-lactamase gene PER-1 was reported to be 68% (36), and in another study by Fallah et al. in 2014 who investigated the prevalence of PER, VEB, IMP and VIM genes in Iran on 108 isolates of Acinetobacter baumanni, the prevalence of these genes was reported as 71, 36, 3, and 15%, respectively (37). According to the current study, the most effective antibiotics for the treatment of UTIs caused by ESBLs-producing Klebsiella strains are Amikacin and Imipenem.
The reasons for the effectiveness of imipenem may be low consumption of this antibiotic in the medical staff, its lack of outpatient use in the community since this antibiotic in Iran is a hospital drug and is not prescribed without indication, as well as the absence or low presence of carbapenemase enzymes in strains related to the family of Enterobacteriaceae (26, 38, 39). The extensive and arbitrary use of cephalosporins has led to the proliferation of ESBLs-producing organisms, which makes these organisms resistant to a wide range of antibiotics from this category. Infections caused by these resistant microorganisms prolong the treatment process and increase the length of the hospitalization and increase the risk of death.
5.1. Conclusions
In this study, although a number of strains were resistant to several drug classes, they lacked the β-lactamase OXA-10 and PER genes. According to numerous studies, the expression of the ESBL phenotype results from the production of broad-spectrum plasmid of group A; therefore, the difference in the prevalence of positive phenotypes and negative genotypes should be sought in the presence of other β-lactamase genes of the group A that cause different outbreaks in different regions.