1. Background
Exointestinal pathogenic Escherichia coli are one of the most important causes of urinary tract infections (UTI). Since the bacteria have a large number of virulence factors and due to the ability to develop rapid resistance to antibiotics, E. coli causing UTI is called uropathogenic E. coli (UPEC). It has been reported that UTI are placed at the top of nosocomial and community-acquired infections. Escherichia coli in community-acquired UTI are reported to be the most common pathogen in Turkey (1, 2).
The most important virulence factor of E. coli that is commonly isolated in UTIs is pyelonephritis-associated P fimbriae (pap). Adhesion was the most important task of P fimbriae. P fimbriae; consists of two subunits, a large subunit, papA and the gene papG. The pap gene cluster consists of 11 genes encoding the foremost component of the pilus rod (papA), which determines 11 different serogroups, and a terminally located adhesion, papG (3, 4).
Antibacterial resistance is a problem faced by the medical world and is getting more and more important every day. Escherichia coli is known to be one of the important beta lactamase producers. Bacteria that produce the beta-lactamase enzyme have a serious resistance mechanism that neutralizes the beta-lactam antibiotics by hydrolysis. At least 350 different beta-lactamase enzymes have been identified. One of the most important enzyme groups are the plasmid encoded extended spectrum beta lactamases (ESBL). Extended spectrum beta lactamases are a group of enzymes that hydrolyze antibiotics, including those containing new cephalosporin. They are found in a significant percentage of E. coli isolates (5). Especially in the last decade, multi-drug resistant E. coli isolates producing ESBLs are an important problem in Turkey as well as in the world (5, 6). Health Protection Agency of Turkey reported that the rate of ESBL-producing E. coli isolates were 21% in 2011 (6).
2. Objectives
This study was aimed at the isolation of E. coli from the urine of patients with UTI using conventional methods, the determination of antibiotic resistance by the disc diffusion test (DDT) and investigation of ESBL enzymes using the double disk diffusion test (DDDT) of isolates; presence of the ESBL genes (blaTEM, blaCTX, blaOXA, blaSHV) and P fimbriae genes (papA, papG) were also investigated using the polymerase chain reaction (PCR).
3. Methods
3.1. Ethics Statement
Ethics Committee of Adnan Menderes University Faculty of Medicine approved the study protocol (413/2014).
3.2. Patients
Uropathogenic E. coli isolates were collected from the Infectious Diseases and Clinical Microbiology Laboratory of Adnan Menderes University Hospital, during a twelve months period in 2015. In the study, 20 - 30 mL of morning sterile urine was taken from patients who had not received antibiotic treatment for at least 15 days as material. Materials were delivered to the lab within latest 20 minutes at the latest.
3.3. Isolation of UPEC
Escherichia coli for the study were isolated from patients with urinary infections hospitalized in Adnan Menderes University Hospital. Collected samples were cultured in blood agar (Merck, Germany) and EMB Agar (Merck, Germany) and incubated for 24 hours at 37°C. Suspicious colonies were chosen and streaked on EMB Agar passaged. Gram staining, oxidase and other biochemical tests (indole, methyl red-voges proskauer, and citrate) were then performed (7). Isolation and identification were performed by conventional biochemical tests, as well as by PCR. We examined the mid-stream clean-catch urine samples of 264 adult patients with the diagnosis of acute uncomplicated UTI. Microorganisms were included in our work when they occurred as a pure culture and at a concentration of more than 105 CFU/ml. The isolates were kept in brain heart infusion broth (Merck, Germany) with 20% glycerol at -20°C.
3.4. Antimicrobial Susceptibility Test and Double Disk Diffusion Test
The Kirby Bauer disc diffusion method was used to detect antibiotic susceptibilities of isolates on Mueller-Hinton agar (Oxoid Ltd, UK) according to recommendations of the Clinical and Laboratory Standards Institute (CLSI) (8). The following Bioanalyse AST (Turkey) discs were used: Amoxicillin-clavulanate (20/10 μg) was placed in the center of the plate with ceftriaxone (30 μg), cefotaxime (30 μg), cefpodoxime (10 μg), aztreonam (30 μg), amikacin (30 μg), gentamicin (10 μg), piperacillin-tazobactam (100/10 μg), ampicillin/sulbactam (10/10 μg), cefazolin (30 μg), cefepime (30 μg), cefuroxime (30 μg), cefoperazone sulbactam (30 μg), imipenem (10 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), colistin (10 μg), nitrofurantoin (300 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), fosfomycin (200 μg). The CLSI recommends that, in DDDT a zone size of ≥ 5 mm difference between the cephalosporin disks should be considered as significant, and indicates ESBL production (8). Escherichia coli ATCC 25922 strains were used as quality control strains in antibiotic susceptibility tests.
3.5. DNA Extraction
We inoculated all isolates aerobically on tryptic soy agar (Merck, Germany) for 18 - 24 hours at 37°C. Genomic bacterial DNA was extracted from bacteria using a commercial DNA extraction kit (InstaGene Matrix, Bio-Rad, USA) according to the manufacturer’s protocol. For this, isolated bacterial colony suspended in 1 ml sterile distilled water. After centrifugation at 10.000 rpm the supernatant was removed. The pellet was suspended in 200 μL of Instagene matrix and vortexed, followed by heating at 56°C for 15 min. The samples were vortexed again and heated at 100°C for 8 min and then centrifuged. The prepared DNA was stored at -20°C and 2 μL supernatant was used template DNA in each PCR.
3.6. Polymerase Chain Reaction
The primers used in this study are listed in Table 1. We performed PCR amplifications in 50 μL reaction mixtures containing final concentrations of 10X Taq buffer solution 1X, 25 mM magnesium chloride (MgCl2) 2 mM, 10 mM dNTP 0.2 mM, 100 pmol primer 0.4 pmol (for each), 5U Taq DNA polymerase (Fermentas) 1.5 and 2 μL of each template DNA. We amplified each gene separately. The DNA was amplified using the following protocol: held at 94ºC for 3 minutes, followed by 35 cycles of denaturation (94ºC for 30 seconds), annealing (44 - 63ºC for 30 seconds) and extension (72ºC for 1 minute), with a single final extension of 7 minutes at 72ºC. Using a 1.5% agarose gel for 45 minutes at 100 V and stained with Safe View (ABM, Canada), we visualized PCR products by electrophoresis. The primers used, together with sequences, amplicon lengths, target genes and sources are shown in Table 1. (9-15).
| Primer | Sequence (5’ - 3’) | Amplicon Length, bp | Target Gene | Source |
|---|---|---|---|---|
| usp-F | CCGATACGCTGCCAATCAG | 884 | usp | 9 |
| usp-R | ACGCAGACCGTAGGCCAGAT | |||
| SHV-F | CGCCTGTGTATTATCTCCCT | 293 | blaSHV | 10 |
| SHV-R | CGAGTAGTCCACCAGATCCT | |||
| CTX-M-F | CGCTGTTGTTAGGAAGTGTG | 569 | blaCTX | 10 |
| CTX-M-R | GGCTGGGTGAAGTAAGTGAC | |||
| TEM-F | ATAAAATTCTTGAAGACGAAA | 1080 | blaTEM | 11 |
| TEM-R | GACAGTTACCAATGCTTAATCA | |||
| OXA-F | ACCAGATTCAACTTTCAA | 598 | blaOXA | 12 |
| OXA-R | TCTTGGCTTTTATGCTTG | |||
| papAH-F | ATGGCAGTGGTGTCTTTTGGTG | 720 | papA | 13 |
| papAH-F | CGTCCCACCATACGTGCTCTTC | |||
| papG allel I-F | TCGTGCTCAGGTCCGGAATTT | 461 | papG allel I | 14 |
| papG allel I-F | TGGCATCCCCCAACATTATCG | |||
| papG allel II-F | GGGATGAGCGGGCCTTTGAT | 190 | papG allel II | 15 |
| papG allel II-R | CGGCGGCCCAAGTAACTCG | |||
| papG allel III-F | GGCCTGCAATGGATTTACCTGG | 258 | papG allel III | 15 |
| papG allel III-R | CCACCAATTGACCATGCCAGAC | |||
| papG allel II/III-F | CTGTAATTACGGAAGTGATTTCTG | 1070 | papG allel II-III | 13 |
| papG allel II/III-R | ACTATCCGGCTCCGGATAAACCAT |
3.7. Statistical Analyses
Statistical analyses were performed by the commercial statistical software SPSS version 21.0 (SPSS Inc., USA). The Chi square test was used to compare the relationship between antibiotic susceptibility and resistance status in ESBL producing and non-producing strains, and in isolates carrying and non-carrying P fimbriae. The results were evaluated at a confidence interval of 95%. A P value < 0.05 was considered as significant.
4. Results
4.1. Isolation and Identification
From 264 urine specimens 133 (133/264: 50.4%) E. coli were isolated and identified using standard biochemical tests. The results were confirmed genotypically. Distribution of infected E. coli isolates according to sex demonstrated that 57.1% (76/133) of the patients were female and 42.9% (57/133) were male.
4.2. Antimicrobial Susceptibility Test and Double Disk Diffusion Test
It was determined that 133 E. coli isolates were 50.3% resistant to ciprofloxacin, 42.1% to trimethoprim sulfamethoxazole, 13.5% to cefazolin, 9.8% to amoxicillin-clavulanic acid and 8.3% to norfloxacin resistant; resistance rates against the other 15 antibiotics used ranged from 5.3% to 0.8% (Table 2).
| Antibiotic | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Ciprofloxacin | 63 (47.4) | 3 (2.3) | 67 (50.3) |
| Trimethoprim-sulfametaxazole | 76 (57.1) | 1 (0.8) | 56 (42.1) |
| Cefazolin | 115 (86.5) | - (0.0) | 18 (13.5) |
| Amoxicillin-clavunic acid | 120 (90.2) | - (0.0) | 13 (9.8) |
| Norfloxacin | 122 (91.7) | - (0.0) | 11 (8.3) |
| Gentamicin | 126 (94.7) | - (0.0) | 7 (5.3) |
| Cefuroxime | 126 (94.7) | - (0.0) | 7 (5.3) |
| Nitrofurantoin | 126 (94.7) | - (0.0) | 7 (5.3) |
| Piperacillin-tazobactam | 119 (89.4) | 7 (5.3) | 7 (5.3) |
| Cefotaxime | 126 (94.7) | - (0.0) | 7 (5.3) |
| Fosfomycin | 127 (95.5) | - (0.0) | 6 (4.5) |
| Cefepime | 127 (95.5) | - (0.0) | 6 (4.5) |
| Cefoperazone-sulbactam | 122 (91.7) | 5 (3.8) | 6 (4.5) |
| Ampicillin-sulbactam | 124 (93.3) | 4 (3.00) | 5 (3.7) |
| Amikacin | 127 (95.4) | 3 (2.3) | 3 (2.3) |
| Cefpodoxime | 130 (97.7) | - (0.0) | 3 (2.3) |
| Ceftriaxone | 129 (97.0) | 1 (0.8) | 3 (2.3) |
| Aztreonam | 132 (99.2) | - (0.0) | 1 (0.8) |
| Colistin | 132 (99.2) | - (0.0) | 1 (0.8) |
| Imipenem | 132 (99.2) | - (0.0) | 1 (0.8) |
aValues are presented as No. (%).
According to DDDT results 49.6% of isolates were found to be ESBL positive while 50.4% were ESBL negative. Of the 66 isolates identified as ESBL positive, 71.1% were ciprofloxacin, 63.6% were trimethoprim-sulfamethoxazole, 10.6% were cefazolin, 9.1% were amoxicillin-clavulanic acid, gentamycin, norfloxacin and piperacillin-tazobactam resistant; resistance rates against the other 13 antibiotics ranged from 6.0% to 1.5% (Table 3).
| Antibiotic | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Ciprofloxacin | 19 (28.9) | - (0.0) | 47 (71.1) |
| Trimethoprim-sulfametaxazole | 22 (33.4) | 2 (0.0) | 42 (63.6) |
| Cefazolin | 59 (89.4) | - (0.0) | 7 (10.6) |
| Amoxicillin-clavunic acid | 60 (90.9) | - (0.0) | 6 (9.1) |
| Gentamicin | 60 (90.9) | - (0.0) | 6 (9.1) |
| Norfloxacin | 60 (90.9) | - (0.0) | 6 (9.1) |
| Piperacillin-tazobactam | 54 (81.2) | 6 (9.1) | 6 (9.1) |
| Cefuroxime | 62 (94.0) | - (0.0) | 4 (6.0) |
| Cefepime | 62 (94.0) | - (0.0) | 4 (6.0) |
| Cefoperazone-sulbactam | 56 (84.9) | 6 (9.1) | 4 (6.0) |
| Cefpodoxime | 63 (95.5) | - (0.0) | 3 (4.5) |
| Ceftriaxone | 63 (95.5) | - (0.0) | 3 (4.5) |
| Fosfomycin | 63 (95.5) | - (0.0) | 3 (4.5) |
| Cefotaxime | 63 (95.5) | - (0.0) | 3 (4.5) |
| Amikacin | 62 (94.0) | 2 (3.0) | 2 (3.0) |
| Aztreonam | 64 (97.0) | - (0.0) | 2 (3.0) |
| Imipenem | 64 (97.0) | - (0.0) | 2 (3.0) |
| Colistin | 65 (98.5) | - (0.0) | 1 (1.5) |
| Ampicillin-sulbactam | 65 (98.5) | - (0.0) | 1 (1.5) |
| Nitrofurantoin | 65 (98.5) | - (0.0) | 1 (1.5) |
aValues are presented as No. (%).
Of the 67 isolates identified as ESBL negative, 29.8% were ciprofloxacin, 23.9% were trimethoprim-sulfamethoxazole, 16.4% were cefazolin, 9.0% were amoxicillin-clavulanic acid and norfloxacin and 8.5% were resistant to nitrofurantoin; the resistance rates of the other 14 antibiotics varied between 6.0% and 0.0% (Table 4).
| Antibiotic | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Ciprofloxacin | 44 (65.7) | 3 (4.5) | 20 (29.8) |
| Trimethoprim-sulfametaxazole | 51 (76.1) | - (0.0) | 16 (23.9) |
| Cefazolin | 56 (83.6) | - (0.0) | 11 (16.4) |
| Amoxicillin-clavunic acid | 61 (91.0) | - (0.0) | 6 (9.0) |
| Norfloxacin | 61 (91.0) | - (0.0) | 6 (9.0) |
| Nitrofurantoin | 61 (91.5) | - (0.0) | 6 (8.5) |
| Cefotaxime | 63 (94.0) | - (0.0) | 4 (6.0) |
| Fosfomycin | 64 (95.5) | - (0.0) | 3 (4.5) |
| Ampicillin-sulbactam | 61 (91.5) | 4 (6.4) | 1 (1.5) |
| Gentamicin | 66 (98.5) | - (0.0) | 1 (1.5) |
| Piperacillin-tazobactam | 66 (98.5) | - (0.0) | 1 (1.5) |
| Cefepime | 66 (98.5) | - (0.0) | 1 (1.5) |
| Ceftriaxone | 66 (98.5) | - (0.0) | 1 (1.5) |
| Amikacin | 66 (98.5) | - (0.0) | 1 (1.5) |
| Cefuroxime | 67 (100.0) | - (0.0) | - (0.0) |
| Cefoperazone-sulbactam | 67 (100.0) | - (0.0) | - (0.0) |
| Cefpodoxime | 67 (100.0) | - (0.0) | - (0.0) |
| Aztreonam | 67 (100.0) | - (0.0) | - (0.0) |
| Imipenem | 67 (100.0) | - (0.0) | - (0.0) |
| Colistin | 67 (100.0) | - (0.0) | - (0.0) |
aValues are presented as No. (%).
4.3. Examination of GSBL Enzyme Types
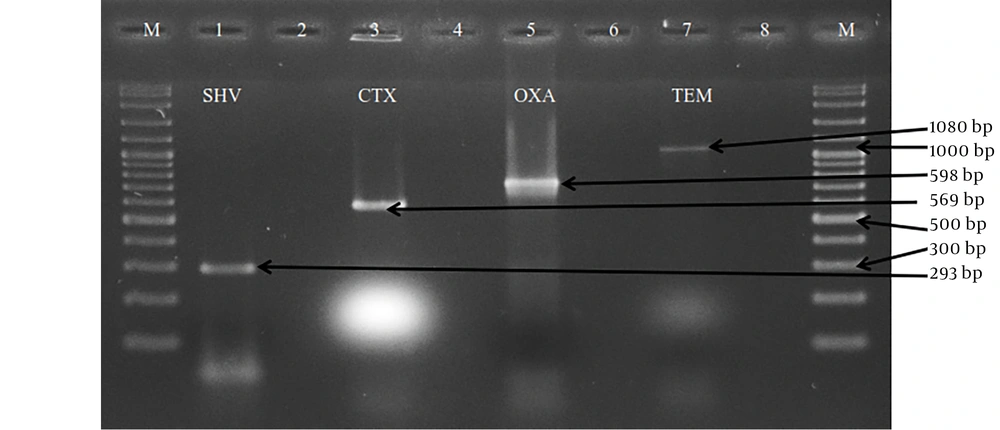
A total of 133 E. coli were examined for their ability to carry the ESBL genes by PZR. TEM, SHV, CTX-M and OXA primers were used for typing the ESBL enzyme. These isolates had four types of ESBL genes. 59.4% (79/133) of the isolates carried at least one of the examined ESBL genes. Extended spectrum beta lactamases genotypes were found among these isolates with the predominance of TEM-type (43/133: 32.3%), followed by CTX-M-type (42/133: 31.6%), OXA-type (28/133: 21.1%) and SHV-type (3/133: 2.3%) (Figure 1).
The gel electrophoresis images of the pap genes. 1: papA positive isolate (720 bp) 3: papG allele I positive isolate (461 bp) 5: papG allele II positive isolate (190 bp) 7: papG allele III positive isolate (258 bp) 9: papG allele II - III positive (1070 bp) 2 - 4 - 6 - 8: negative control (mastermix without DNA) M: 100 bp DNA ladder (Vivantis®)
4.4. Examination of P Fimbriae Genes (papA and papG)
A total of 51 pap (papA + papG) genes were detected in 27.0% (36/133) of isolates; 12.0% (16/133) of them were found to be papA while 26.3% (35/133) were found to be papG. papG allele genotypes were found among these isolates with the predominance of allele II (33/133: 24.8%), followed by allele III (7/133: 5.3%) and allele I (1/133: 0.8%). Allele II and III were present in 26.3% (35/133) of the isolates.
4.5. Statistical Analyses
As a result of the Chi-square test conducted to compare antibiotic susceptibility and resistance status, (1) it was detected that ciprofloxacin and trimethoprim-sulfamethoxazole antibiotics were significantly different between the groups, with regards to ESBL-producing and nonproducing isolates, (2) it was detected that there was a significant difference between the ciprofloxacin resistance and papG presence in the groups, with regards to P fimbriae carrying and non-carrying isolates (P < 0.05).
5. Discussion
Escherichia coli isolates from human clinical specimens especially UTI cause significant public health problems because the majority of these strains carry multidrug resistance genes (16). However, mobile genetic elements for example plasmids can spread ESBL-related genes via horizontal gene transfer in other Gram-negative bacteria (17). The vast majority of UTIs are caused by a single species of bacteria, and E. coli have been reported as the most frequently isolated infectious agent from UTI in many studies conducted in Turkey and around the world. In this study, E. coli was isolated in a high proportion (50.4%) of UTIs, similar to the above studies; a large portion of these isolates (57.1%) were obtained from female patients. This is consistent with previous findings which report that UTI is more common in females (2, 6, 16, 17). This difference could be due to anatomic differences, hormonal effects or several clinical factors.
It is important to have knowledge about the resistance to antimicrobials at the country or even region level during UTI treatment. As a result, there is always a need for studies to determine the susceptibility profiles of agents. Numerous studies have been carried out on the antibiotic resistance of UPECs in Turkey and in the world and continue to be performed. The most commonly used drugs in the management of UTI are trimethoprim-sulfamethoxazole and quinolones. In the study, E. coli isolates were found to be most resistant to ciprofloxacin, trimethoprim sulfamethoxazole, cefazolin, amoxicillin-clavulanic acid and norfloxacin, which are the most frequently, used antibiotics in empirical therapy. These findings have also been reported in other studies (18, 19). It is recommended to use other antibiotics such as imipenem, aztreonam, colistin in the treatment. As a result, these antibiotics, which are now less resistant, can be used for the treatment of UTI’s. This may be due to increased use of these antibiotics, and resistance transfer (17).
A different problem is the increased occurrence of ESBL producing organisms causing UTI, and the emergence of severe health problems due to antibiotic resistance. Extended spectrum beta lactamases can be transferred to other strains via plasmid and transposon, further increasing the significance of this ESBL-resistant isolates (3, 15, 19). Senbayrak et al. reported a significant increase in ESBL prevalence from 12.5% to 44.7% in both male and female hospitalized patients, between 2004 - 2012 (20). Toka Ozer et al. reported the rate of ESBL production in E. coli strains was 13.1% in 2018 (1). Antibiotic resistance rates have also been reported to increase for ESBL producing E. coli. These rates were higher than 70% for fluoroquinolones, amoxicillin-clavulanic acid and trimethoprim-sulfamethoxazole. The prevalence of ESBL-producing E. coli for inpatients in Switzerland is 6.6% (21), 10.4% in Saudi Arabia (22) and 52.9% in China (23). In our study, 49.7% of the isolates were phenotypically ESBL positive. Comparing antibiotic resistance profiles of ESBL producing and non-ESBL producing E. coli strains, there is great similarity in the order of resistant antibiotics, although there is a difference in resistance rates. Resistance rates were found to be higher in the presence of ESBL.
In ESBL-producing E. coli, resistance to different antibiotics is often observed because the coding genes are usually in the same plasmids (24). According to results of this study, 59.4% of the isolated bacteria carried at least one of the examined ESBL genes. The percentages of genes detected were 32.3%, 31.6%, 21.1%, 2.3% for blaTEM, blaCTX, blaOXA, blaSHV, respectively. Recent studies have unveiled the majority of blaCTX among ESBL positive phenotype all over the world. In these studies, the OXA gene was also not detected (25, 26). Mohajeri et al. in Iran demonstrated that 93.3%, 68.2%, 43.2%, 31.8%, and 22.7% of CTX-M, SHV, TEM, OXAI and OXAII genes were present in UPEC (27). In our study, the TEM gene was shown to be of a high frequency compared to CTX, OXA and SHV genes. However, further studies are required to identify other genes in ESBLs producing E. coli isolates.
P fimbriae are common virulence factors of UPEC, which play an important role in the pathogenesis of ascending UTIs and pyelonephritis in humans (2, 5). P fimbriae consist of heteropolymeric fibres of different protein subunits, encoded by the papA-K gene operon (28). papA is an important structural subunit of the pilus associated with pyelonephritis, and has critical prescription for pilus synthesis. papG is Gal(1-4)Gal-specific pilus tip adhesin molecule. papG has three alleles. Allele I is known as J96-associated, allele II is known as pyelonephritis-associated, while allele III is known as cystitis-associated papG variant (5, 28). Of our isolates, 27% (36 of them) carried a total of 51 genes (papA or papG). In our isolates, 26.3% of papG was detected while 12% of papA was detected. A study from Ankara in Turkey, parallel to our study, revealed that almost 23% E. coli isolates were positive for pap genes (29). In Iran, Mohajeri (30), Farshad and Emamghorashi (31) and Rahdar et al. (32) reported the frequencies of pap genes as 20.5%, 30.2%, 57%, respectively. These differences may be due to the differences in geographical regions. The pap gene encodes for P-fimbriae that allows bacteria to adhere to epithelial surfaces and protect them against urine lavage, thus allowing them to ascend further to even cause serious disease (30).
It was reported that was extended spectrum β-lactamase producing E. coli infections alarmingly increased in recent years in Turkey (1, 20). On the other hand, inappropriate and excessive use of antibiotics has resulted drug resistance. This limits treatment options. However, because of the rates of antibiotic resistance between regions are very variable, it is important to know the resistance profile of the region in the selection of antibiotics to be preferred in empirical treatment. When regional differences in antibiotic resistance rates are taken into consideration, antibacterial resistance should be regularly monitored in each geographical area in order to determine appropriate treatment strategies and reduce treatment costs.
5.1. Conclusions
Extended spectrum -lactamase producing E. coli infections associated with UTI increased in last years in Turkey. Further studies should be designed to investigate plasmids of ESBL positive E. coli stains isolated from UTI and whether plasmids are responsible for ESBL production