1. Background
The prevalence of fungal infections caused by yeasts and yeast-like species, particularly the genus of Candida, significantly increased due to extensive application of immunosuppressive drugs, predisposing factors, and particular conditions. Some rare and uncommon yeast species emerged as a cause of outbreak with high mortality or mixed infections non-responsive to routine antifungals (1, 2). Clinical manifestations of yeasts and yeast-like infections ranging from mild superficial to systemic infections are frequently and usually benign in immunocompetent individuals (3). Candida species are the major cause of yeast infections and Candida albicans is still the most frequently isolated yeast pathogen (4).
Non-albicans Candida species, e g, C. tropicalis, C. parapsilosis, C. krusei, C. glabrata and some uncommon yeast species such as Trichosporon and more specifically C. auris complex (C. auris, C. haemulonii, C. pseudohaemulonii, C. duobushaemulonii, and C. haemulonii var. vulnera) with a reduced susceptibility to antifungal agents become a serious clinical challenge and thus the isolates need to be properly identified (1, 2, 5-7). Candida auris and its closely related C. haemulonii emerged as a causative agent of invasive candidiasis that were extensively drug-resistant in hospital outbreaks (8, 9). Candida auris is often misidentified with C. haemulonii by phenotypic methods. Due to the high degree of similarity between C. auris complex, identification is problematic. Morphological and physiological approaches to identify the species are time-consuming and frequently unspecific (6, 10).
2. Objectives
The current study was aimed to determining the prevalence of common and uncommon yeasts particularly Candida species isolated from patients with candidiasis referred to medical mycology labs, and identifying the species using molecular methods that cannot be readily differentiated with phenotypic assays.
3. Methods
3.1. Ethics Statement
The current study was approved by the Ethics Committee of Fasa University of Medical Sciences (ethical code: 93210/D,97,247016), Fasa, Iran, and written informed consent was obtained from the patients.
3.2. Yeast Isolates
Totally, 1200 clinical samples were collected from patients suspected of fungal infection (age range 1 - 84 years) referring to the medical mycology laboratory, from 2012 to 2014 as part of routine medical inquiry. In total, 110 yeast and yeast-like species were isolated from the nail (n = 46), cutaneous (n = 37), bronchoalveolar lavage (BAL) (n = 9), sputum (n = 7), mucosal specimens (n = 3), vagina (n = 2), ear discharge (n = 1), and urine (n = 1) samples. Isolates were initially examined for clinical specimens by direct microscopic examination (10% KOH), afterward cultured on malt-extract agar (Difco, USA) and incubated at 35°C for two days. The grown yeasts within 48 hours were initially identified by germ-tube test, and CHROMagar Candida medium (CHROMagar Company, Paris, France) as previously described (11).
3.3. Molecular Identification
By glass bead disruption method, genomic DNA was extracted as previously described and stored at -20°C prior to use (12). The PCR-RFLP method was conducted as previously explained (1). In a nutshell, PCR amplification of ITS1-5.8S-ITS2 rDNA regions was carried out in a final volume of 25 µL using the primers ITS1 (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3’) (Metabion, Germany) (13). Each reaction consists of 1 µL template DNA, 1 µL of each primer at 10 pmol, 12.5 µL MasterMix (Ampliqon, Denmark) containing 5 mM dNTP, 0.5 U Taq DNA polymerase, and 10X PCR buffer and 9.5 µL double-distilled water. PCR conditions were adjusted with cycles of five minutes at 94°C for primary denaturation, followed by 35 cycles at 94°C (30 seconds), 55°C (45 seconds) and 72°C (60 seconds), and final extension at 72°C for seven minutes. Subsequently, PCR products were digested in a final reaction volume of 30 μL containing 17 μL water, 2 μL of 10X FastDigest buffer, 1 U of FastDigest restriction enzyme MspI (Fermentas, Thermo Fisher Scientific Inc., USA) and 10 μL PCR product at 37°C for 10 minutes. After the digestion with MspI enzyme, the PCR products were evaluated based on the molecular sizes by gel electrophoresis for Candida species according to Mohammadi et al. (1). Candida albicans CBS 562, C. krusei ATCC 6258, and C. parapsilosis ATCC 22019 were used as standard strains in the study.
In addition, the partial amplification of the hyphal wall protein 1 (hwp1) gene was used to differentiate C. albicans species complex (C. africana, C. albicans, C. dubliniensis, and C. stellatoidea) as described by Romeo and Criseo (14). Briefly, PCR amplification of hwp1 gene was performed using the forward (5’-GCT ACC ACT TCA GAA TCA TCA TC-3’) and reverse primers (5’-GCA CCT TCA GTC GTA GAG ACG-3’) (Metabion, Germany) by producing three different DNA fragments as follows: 941 bp for C. albicans, ~740 bp for C. africana, 569 bp for C. dubliniensis, and ~800 bp for C. stellatoidea type I (14, 15). Amplified and digested products were stained with KBC power load (Kawsar Biotech Co., Iran) and run on 1.5% and 2% agarose gel electrophoresis in TBE buffer (0.09 M Tris, 0.09 M boric acid, and 2 mM EDTA; pH 8.3), respectively, and visualized by gel document device (12). Candida albicans CBS 562 and C. africana CBS 8781 were applied as standard strains in the study. For undifferentiated species by PCR-RFLP, sequencing was performed on an ABI 3730xl automatic sequencer (Applied Biosystems, Foster City, CA) using the primers ITS1 and ITS4, and for confirmation C. africana hwp1 gene was amplified. Sequence data were obtained and compared with GenBank database using the basic local alignment search tool (BLAST) (http://www.ncbi.nlm.nih.gov). All obtained sequences were deposited in GenBank under the accession numbers KY091885, KY112734-KY112744, and KY238037-KY238038.
4. Results
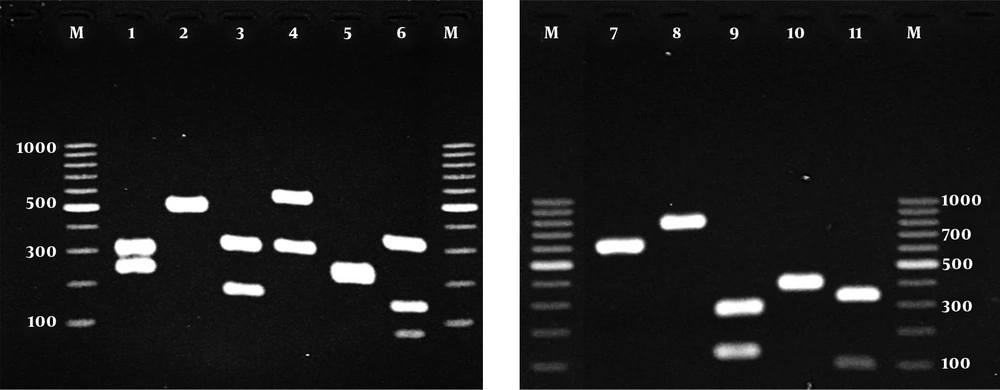
Table 1 shows distribution of yeast species based on the site of infection in patients referring to the Mycology Lab of Baghban Clinic, Sari, Iran. In the current study, of 1200 clinical samples, 110 (9.16 %) were positive for yeast and yeast-like infections, of which 70 (63.6%) samples were obtained from females and 40 (36.4%) from males. Most of the samples were attributed to nail (41.8%), followed by skin (32.7%) and BAL (8.2%). Thirty-seven (80.4%) yeast species isolated from nail samples belonged to females. As shown in Table 2, depending on the species, the size of ITS rDNA region before and after endonuclease digestion with MspI, amplification of the ITS region in all yeast isolates yielded the band of approximately 389 to 881 bp.
| Specimen | Nail | Skin | BAL | Sputum | Mouth | Urine | Vagina | Mucosa | Ear Discharge | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Candida albicans | 20 | 8 | 7 | 5 | 4 | 1 | 1 | 46 | ||
| C. parapsilosis | 10 | 6 | 1 | 17 | ||||||
| C. tropicalis | 7 | 5 | 1 | 13 | ||||||
| C. guilliermondii | 7 | 5 | 12 | |||||||
| C. glabrata | 1 | 1 | 1 | 1 | 4 | |||||
| C. krusei | 1 | 2 | 3 | |||||||
| C. famata | 3 | 3 | ||||||||
| C. kefyr | 1 | 1 | 2 | |||||||
| C. haemulonii | 1 | 1 | 2 | |||||||
| Cutaneotrichosporon jirovecii | 2 | 2 | ||||||||
| C. intermedia | 1 | 1 | ||||||||
| C. sorbosivorans | 1 | 1 | ||||||||
| C. africana | 1 | 1 | ||||||||
| C. stellatoidea | 1 | 1 | ||||||||
| Pichia kudriavzevii | 1 | 1 | ||||||||
| Trichosporon asahii | 1 | 1 | ||||||||
| Total | 46 | 36 | 9 | 7 | 5 | 1 | 2 | 3 | 1 | 110 |
| Yeast Species | Size of ITS | Size of MspI-RFLP Fragment |
|---|---|---|
| Candida albicans | 537 | 239, 298 |
| C. parapsilosis | 530 | 530 |
| C. tropicalis | 526 | 186, 340 |
| C. glabrata | 881 | 320, 561 |
| C. krusei | 510 | 250, 260 |
| C. guilliermondii | 607 | 82, 155, 370 |
| C. famata | 639 | 639 |
| C. kefyr | 720 | 720 |
| C. intermedia | 389 | 122, 267 |
| C. haemulonii | ~400 | ~400 |
| C. sorbosivorans | ~440 | ~100, ~340 |
Figure 1 showed the patterns of ITS PCR-RFLP for yeast isolates after digestion with MspI. The accurate identification of the strains remained undifferentiated based on the results of sequencing method.
Agarose gel electrophoresis of ITS-PCR products of yeasts species after digestion with MspI; lanes 1 - 11: C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, C. krusei, C. guilliermondii, C. famata, C. kefyr, C. intermedia, C. haemulonii and C. sorbosivorans, respectively; Lane M: a 100-bp DNA size marker
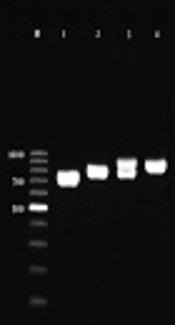
Figure 2 demonstrates the patterns of PCR amplification of hwp1 gene for C. albicans complex isolates. Sequencing results from C. africana strain showed 99% similarity with those of GenBank database using BLAST. There were two different patterns of PCR amplification of hwp1 gene for C. albicans (Figure 2; lanes 3 and 4), which Lane 3 is an atypical isolate of C. albicans, since they exhibited heterogeneous genetic characteristics (14).
Based on the above mentioned molecular methods, C. albicans was the most frequently isolated species (n = 46; 41.8%) followed by C. parapsilosis complex (n = 17; 15.4%), C. tropicalis (n = 13; 11.8%), C. guilliermondii (n = 12; 10.9%), C. glabrata (n = 4; 3.6%), C. krusei and C. famata (n = 3 each; 2.7%), C. kefyr, C. haemulonii and Cutaneotrichosporon jirovecii (n = 2 each; 1.8%) and C. stellatoidea, C. intermedia, C. sorbosivorans, C. africana, Pichia kudriavzevii (teleomorph of C. krusei), and Trichosporon asahii (n = 1 each; 0.9%). As shown in Table 1, several uncommon and less reported yeasts such as C. africana and T. asahii isolated from inguinal region, P. kudriavzevii from stomach mucosa, and two C. jirovecii from cutaneous samples were identified; furthermore, two cases of C. haemulonii (KY112737 and KY112738) and C. sorbosivorans (KY112736) were reported for the first time in Iran. It is noteworthy that C. haemulonii (S49AF, accession number: KY112738) strain was reconfirmed by a novel multiplex end-point PCR as well (16).
5. Discussion
Among hundreds of yeast and yeast-like species, only a few are the normal flora of skin and mucous membranes. These yeasts may cause opportunistic infections in immunocompromised patients under particular conditions (17). In epidemiological studies, most of the yeast isolates belong to Candida species, especially C. albicans (1, 4, 5, 17, 18). Previous studies in different regions of Iran show that Candida species were isolated from various clinical samples rather than other yeasts (2, 5, 18-23). As already mentioned in most studies, among Candida species, C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, and C. krusei were the most common causes of candidiasis in Iran. Several studies showed that Candida onychomycosis was the predominant yeast infection (2, 5, 19, 21), consistent with the current study results (41.8%). In the current study, the most prevalent species isolated from clinical samples was C. albicans, followed by C. parapsilosis, C. tropicalis, and C. guilliermondii. Moreover, some uncommon yeasts including C. africana, T. asahii, and C. jirovecii isolated in the current study that were less reported from Iran so far (24-27).
In recent years, there are some reports of infections caused by uncommon yeasts and yeast-like fungi in the world. For instance, Kubica et al., reported the isolation of 30 T. asahii strains from clinical samples in Brazil (28). Messias Silvestre et al., isolated 112 Trichosporon spp. from perigenital skin with predominance of T. cutaneum, and 30 Trichosporon species strains from urine samples and catheters, mainly T. asahii (29). Chagas-Neto et al., recovered 22 isolates from blood cultures, mostly T. asahii (30). A study from Italy reported 17 cases of invasive infections caused by Trichosporon species (31). Almeida Junior et al., comprehensively reviewed 203 cases of invasive Trichosporon infection in the literature reported from 1994 to 2015 indicating that T. asahii was the predominant species (46.7%) caused trichosporonosis (7). Moreover, Karimi et al. (32), identified 23 uncommon yeasts out of 855 (5.7%) yeast strains isolated from Iran; yeast species not reported in Iran so far (e g, C. magnolia) are closely related with C. sorbosivorans in terms of phylogenic relationships (33). In the current study, C. sorbosivorans (KY112736) was isolated from inguinal region of a 55-year-old female. To the best of our knowledge, it was the first report of C. sorbosivorans as a causative agent of candidiasis. Candida sorbosivorans isolated from contaminated industrial material for the first time by James et al. (33).
As well as, for the first time C. haemulonii (KY112737-38) was reported from Iran, from a 36-year-old male with cutaneous candidiasis and the sputum of a 75-year-old male. This species reconfirmed thereinafter using a novel multiplex (16). Candida haemulonii and its close relative C. auris caused remarkably high mortality, especially in patients with candidemia (8, 9). Candida haemulonii as a member of C. auris complex emerged as an opportunistic fungal pathogen associated with candidiasis. Some species related to C. auris complex were responsible for candidemia and showed antifungal resistance profiles (10). Other members of C. auris complex include C. pseudohaemulonii, C. duobushaemulonii, C. haemulonii var. vulnera, and C. auris (10, 34-37). In most studies, antifungal susceptibility tests of C. auris complex show high resistance to azole derivatives, especially fluconazole; but echinocandins are highly active against C. auris complex (10, 36, 38-40). However, strains resistant to this new class of antifungal agents were also reported (41, 42).
5.1. Conclusions
With the growing population at risk for fungal infections and the emergence of some less virulent or non-pathogenic and uncommon yeasts not readily distinguishable with phenotypic assays, the accurate identification of such microorganisms using molecular methods are warranted.