1. Background
Influenza viruses are one of the most common viruses spreading among the human population worldwide (1). Influenza viruses belong to Orthomyxoviridae type A and type B, which cause diseases in humans. Both types of viruses are responsible for the seasonal influenza epidemics; nevertheless, type B causes milder symptoms and is less frequent (2). The predominant hosts for influenza type B viruses are humans, whereas the primary source for influenza type A viruses are birds. It should be noted that these types of viruses may also spread in other species such as pigs, horses, and bats (3).
Influenza type A viruses are divided into subtypes based on hemagglutinin and neuraminidase surficial glycoproteins. Due to their exposure to the surface of virus, these glycoproteins are the main targets for the host protective immune responses. These can change as a result of antigenic drift, that is, mutations within the gene over time and as a result of antigenic shift (two strains assort again to make a new mixture of surface antigens) (4, 5).
Hence, the vaccine seed has to be changed annually to adjust to the variations in the antigenic sites of viruses, and vaccine development and production take months (6, 7). Conventional vaccines for influenza viruses are based on the major glycoprotein of viruses or their hemagglutinin content. However, the conventional vaccines have crucial drawbacks, the most important of which is the uncertainty in the selection of virus strains (4, 8). Considering the limitations of conventional influenza vaccines, researchers suggested an entirely different approach based on the highly conserved extracellular domain of the viral M2 protein (M2e) (7-9).
The genome of the influenza A virus contains eight segments of negative sense RNA inside the lipid bilayer envelope. Matrix protein 2 (M2) is an influenza A virus structural protein, which serves a crucial function in the virus life cycle. The M2 is a type III membrane protein with 96 amino acid residues (10). The N-terminal ectodomain (M2e) contains 23 residues; this sequence has stayed nearly unchanged since the first human influenza strain was isolated in 1933 (8, 11). As the M2e sequence is extremely conservative, many M2e vaccines have been designed and successfully tested for efficacy against a panel of divergent influenza viruses in animal models (7, 8, 11-13). Due to the weak immunogenic properties of M2e, different approaches have been introduced to link M2e to carriers such as molecular adjuvants. Such adjuvants can be presented in a much more immunogenic form to enhance the immunogenicity of M2e (14).
The adjuvant properties of flagellin (encoded by FliC gene) of Salmonella typhimurium have been demonstrated. FliC is a potent T-cell antigen and can be potentially used as a vaccine adjuvant. Unlike other toll-like receptors (TLR) agonists, flagellin tends to produce mixed Th1 and Th2 responses rather than strong Th1 responses (9, 15). Influenza virus has a large impact on public health. Due to permanent mutations in genome of the virus and the perpetual possibility of producing new viruses that occur as seasonal or pandemic flu, the development of a universal vaccine for this virus is very important.
2. Objectives
The objective of the present study was to have a complete bioimmunoinformatics analysis of M2e.FliC fusion peptide and to design a clone to express it efficiently in Escherichia coli. This fusion peptide may be referred to as a universal vaccine for the human influenza A.
3. Methods
3.1. Bioinformatic Investigations
The first step of the procedure was the analysis of 3M2e (three tandem repeats of M2e sequence), FliC, and 3M2e-FliC amino acid sequences, and then translating these sequences to create the final structure of the protein. The reference sequence of M2e conserved as an epitopic sequence is SLLTEVETPIRNEWGSRSNDSSD, available at http://www.iedb.org with “142017” identification code. The EVETPIRN sequence was detected by anti-M2e specific antibody; thus, by removing this sequence, the epitopic sequence of M2e cannot be recognized by specific antibody. The M2e.FliC fusion construction was translated by the online translating tool of ExPASy server, and the amino acid sequence of the M2e.FliC protein is demonstrated in Figure 1.
PDB ID of 4N8C and MMDB ID of 124024 represent the crystal structure of M2e in complex with the Fab of a protective M2e-specific monoclonal antibody (10, 16). NCBI Reference Sequence of FliC is “WP_003120600.1”, which is available at www.ncbi.nlm.nih.gov/genbank/. The deduced 3M2e.FliC fusion peptide was subjected to protein identification, and the analysis tools on the ExPASy server for the determination of chemical and physical properties are available at http://www.expasy.org/proteomics. Linear epitope prediction and superficial amino acids prediction of 3M2e.FliC were performed by Immune Epitope Database (IEDB) analysis resource. The IEDB analysis resource tools are recommended for B cell epitope mapping and in silico epitope prediction available at http://tools.iedb.org/bcell/ (17). We also employed YASPIN secondary structure prediction that is available at http://www.ibi.vu.nl/programs/yaspinwww/ in order to predict the secondary structure of 3M2e.FliC recombinant protein (18).
Three-dimensional structure of a protein provides information on biological function of protein, its relationship with other components (e.g., ligands, proteins, and DNA), and the phenotypic effects of mutations (19, 20). Hence, many models have been designed based on the two major modeling methods, AB initio and Homology modeling. AB initio modeling method was applied to make 3M2e three-dimensional structure using QUARK server, available at http://zhanglab.ccmb.med.umich.edu/QUARK/. The Ab initio method is used for 3M2e modeling instead of homology modeling method, because there is no model that truly shows the M2e 3D structure. The homology modeling method was employed for the modeling of FliC by applying Raptor-X server (http://raptorx.uchicago.edu/), Phyre2 server (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index), I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), and Modeller v9.15 software (19-22).
Validation of all models was evaluated by Q-mean server (https://swissmodel.expasy.org/qmean/); the output is a score between zero with higher scores showing better quality of the model. Modeling of 3M2e.FliC was performed the same way as FliC modeling. Then, models for FliC and 3M2e.FliC were created using Modeller software by combining the best models achieved from the servers and Modeller software. In fact, some parts of the best-made models were used as templates for building the new model. The best created model was evaluated by Ramachandran plot using PROCESS server (http://www.prosess.ca/) (19). Snap-Gene and Gene-Runner software programs were used for the primer design.
3.2. Experimental Investigations
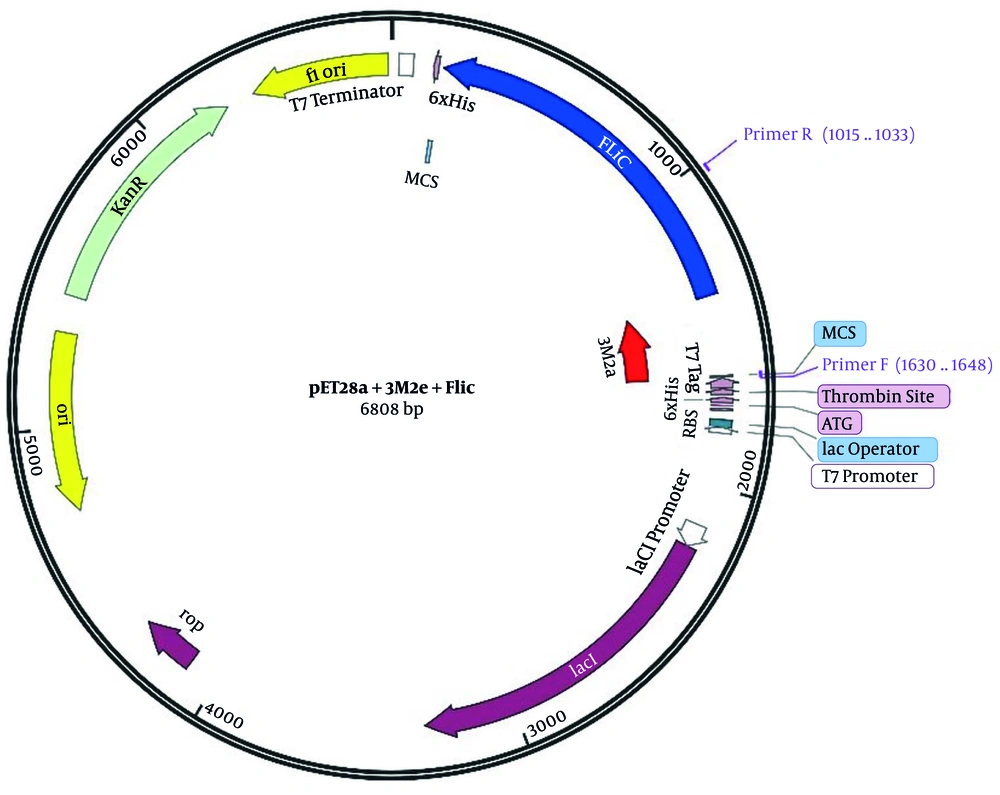
The FliC full-length gene cloned into pET28a through BamHI/HindIII sites (pET28a+Flic) was gifted by Dr. Mahdavi (Pasteur Institute of Iran). Synthesized three tandem repeat sequences of M2e (3M2e) cloned by BamHI sites into pET28a vector (pET28a+3M2e) were also received as a gift from Dr. Fotouhie (Pasteur Institute of Iran). Bioneer kit (cat#K-3030) was used for plasmid purification and 10 mL of Luria-Bertani (LB) agar medium for each purification column. After the purification of pET28a+3M2e plasmid, it was cut by BamHI Fermentas enzyme (cat# ER0055), kept for 3 h at 37°C with 1x TangoTM buffer, and cloned into BamHI linearized pET28a+FliC. Employing this strategy, the 3M2e segment was placed at the upstream of the FliC gene in frame and in fusion with His-tag. Figure 2 demonstrates a schismatic diagram of “pET28a+3M2e.FliC” map.
The correct orientation of “pET28a+3M2e.FliC” was confirmed by PCR. A pair of primers was designed such that the first half of forward primer (GGTCGCGGATCCAGTCTTC) was complementary of the vector and its second half was complementary of the beginning of 3M2e fragment. The reverse primer (GCTTCACTTCGCCGTTCAG) was complementary of the midst of the FliC gene. The pET28a+3M2e-FliC was transformed into the Escherichia coli BL21 (DE3) and ER2566 (Novagen) competent cells. Single colonies from transformed cells were used for the inoculation of 5 mL pre-culture medium containing Kanamycine. The cultures were incubated for 12 h at 37°C and shaken at 180 rpm. Also, 200 µL of the pre-cultures was used for inoculation of 20 mL of LB medium at 37°C to reach the OD600 of 0.8. For protein expression, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the final concentration of 1 mM. The expression was continued at 37°C for 24 h and cells were harvested within the desired intervals (four samples hour by hour and one sample 20 hours after induction). The total cell protein (TCP) was analyzed on 12% SDS-PAGE. The relative expression level was quantified by GelAnalyzer v2010a software.
For further characterization, the separated proteins on SDS-PAGE were electrotransferred to nitrocellulose membrane and western blotting was carried out using Anti-His6 (23) as the primary antibody and anti-mouse IgG peroxidase conjugate (Sigma, USA) as the secondary one. The nitrocellulose sheets were developed by diaminobenzidine (DAB) substrate (23).
4. Results
4.1. Immunoinformatics Analysis Results
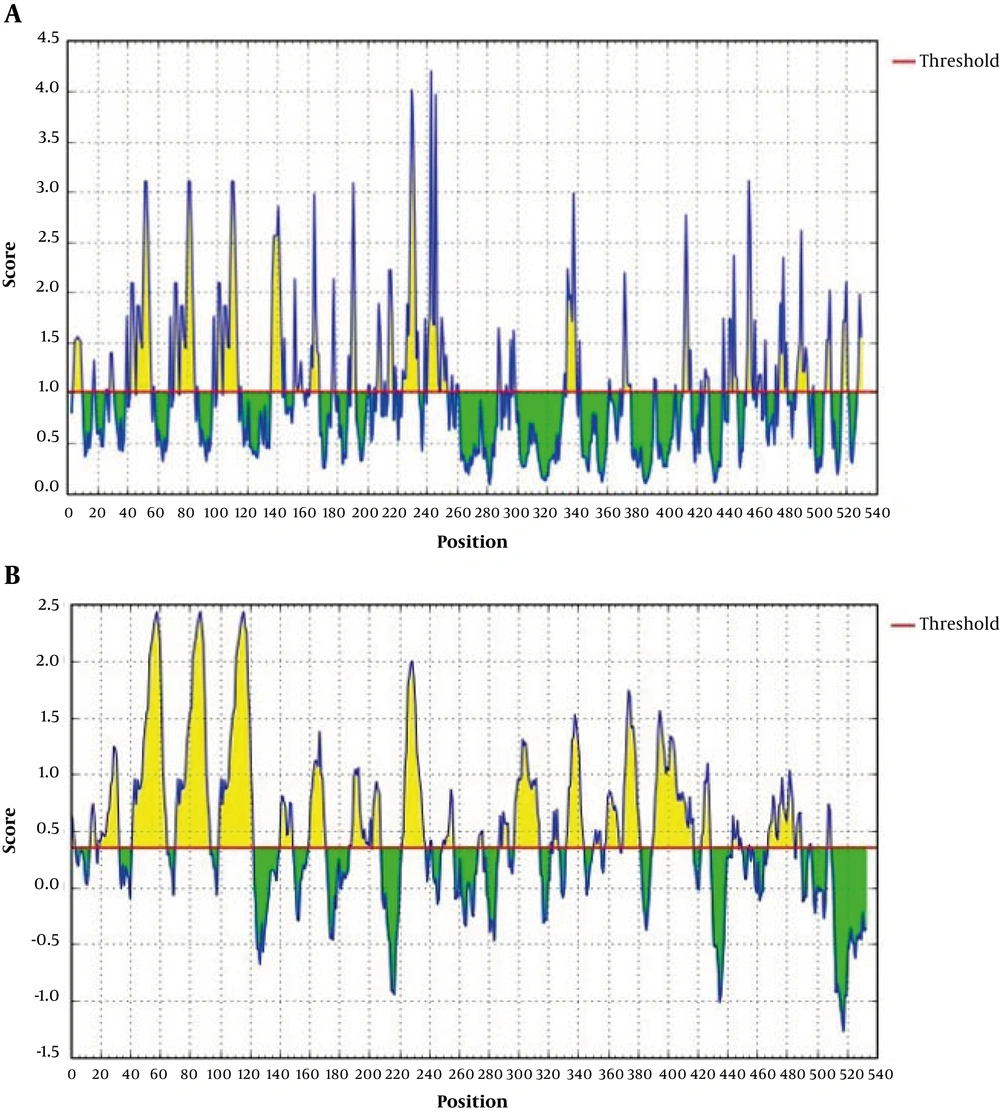
Protein identification and analysis tools on the ExPASy server determined the chemical and physical properties of 3M2e.FliC. The results of this analysis indicated that the number of amino acids was 533 with molecular weight of 54666.8 dalton and theoretical pI of 5.74. At the 3M2e.FliC protein, the total number of negatively charged residues (Asp + Glu) was 54 and the total number of positively charged residues (Arg + Lys) was 44. Based on the analysis by protein identification and analysis tools on the ExPASy server, the estimated half-life of protein is 30 hours for mammalian reticulocytes in vitro, more than 20 hours for the yeast in vivo, and over 10 hours for E. coli in vivo. The instability index (II) was computed at 27.79 confirming the stability of protein. Linear epitope and superficial amino acids prediction of 3M2e.FliC fused construction was performed by IEDB analysis resource. The results of the linear epitope prediction of 3M2e.FliC fused construction and superficial amino acids were predicted using the IEDB analysis resource shown in Figure 3.
A, prediction of 3M2e.FliC linear epitopes generated by IEDB analysis resource. The red line shows the threshold (default 0.35). The residues with scores above the threshold, shown in yellow, are predicted to be part of an epitope. B, prediction of the 3M2e.FliC superficial amino acids generated by IEDB analysis resource. Being a superficial amino acid, a hexapeptide sequence of amino acids must obtain a score greater than 1.0. In this diagram, the superficial amino acids have been shown in yellow above the threshold lane.
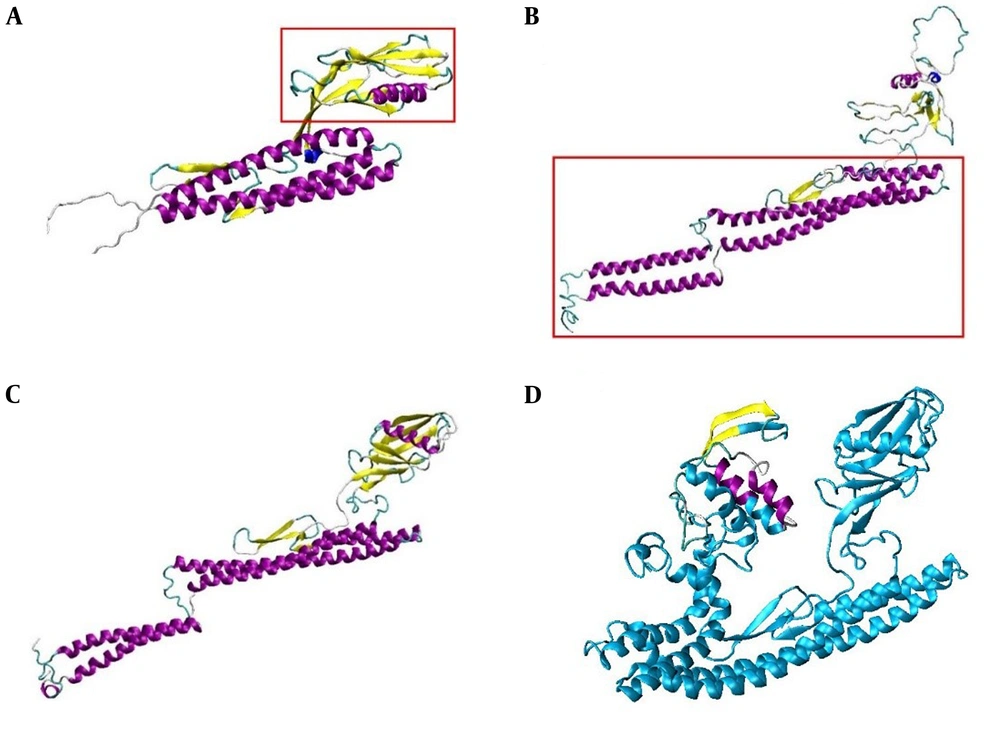
Protein three-dimensional model of 3M2e was constructed. The Ab initio modeling results indicated that Q-mean score of the best model of 3M2e constructed using QUARK server was 0.334. Afterwards, FliC model was made by three servers (i.e., Raptor X, Phyre2, and I-TASSER) and Homology modeling by Modeller v9.15 software. Then, the best model was selected based on Q-mean score. The model provided by I-TASSER server was the best with 0.384 Q-mean score. The 3D-refine server was used in order to increase the quality and Q-mean score. The new score was 0.427. Then, models were made by Modeller v9.15 software and I-TASSER server was selected as template for Modeller v9.15 software to make a new model. The parts marked with red rectangles in Figure 4A and 4B were used as templates to create a new model with Modeller v9.15 software. The model made with this method gained 0.393 Q-mean score. After refining this model with 3D-refine server, it obtained Q-mean score of 0.456. The combinatorial model of FliC is shown in Figure 4C.
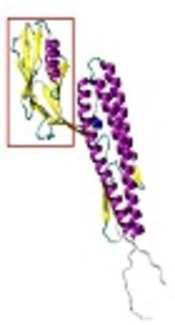
A, FliC modeling with Modeller v9.15. Alpha helices and beta sheets are shown by violet and yellow color, respectively. The part in red rectangle was chosen as template for modeling of FliC with Modeller v9.15. B, FliC modeling with I-TASSER server. Alpha helices and beta sheets are shown by violet and yellow color, respectively. The part in red rectangle was chosen as template for modeling of FliC with Modeller v9.15. C, combinatorial model of FliC made by Modeller v9.15 software. The best models, made by Modeller v9.15 and I-TASSER server, were selected as templates for making this model. Alpha helices and beta sheets are shown in violet and yellow, recpectivly. D, the combinatorial model of 3M2e.FliC was made by Modeller v9.15 software; 3M2e epitopes are shown in yellow and violet.
According to Ramachandran diagram and its analysis, six amino acids from the total 413 amino acids of the FliC combinatorial model had inappropriate Psi and Phi angles. Finally, 3M2e.FliC model was made with two servers (i.e., Phyre2 and I-TASSER) and Modeller v9.15 software on the basis of homology modeling. Q-mean score for the best model constructed by Phyre2 server was 0.325, Q-mean score for the best model of I-TASSER server was 0.255, and the score for the best model made by Modeller v9.15 software was 0.349. The constructed model by Modeller 9.15 software was refined with the 3D-refine server and the new score was 0.404. Then, the constructed 3M2e model and the combinatorial model of FliC were merged and a 3M2e.Flic combinatorial model was produced by Modeller v9.15 software (Figure 4D). After refining the model with 3D-refine server, 0.422 Q-mean score was obtained. The model clearly indicated that M2e epitopes are accessible on the protein surface. According to Ramachandran diagram, 13 amino acids from 533 amino acids of the 3M2e.FliC combinatorial model had inappropriate Psi and Phi angles; therefore, they displayed they are outside the standard range of Ramachandran diagram.
4.2. Experimental Results
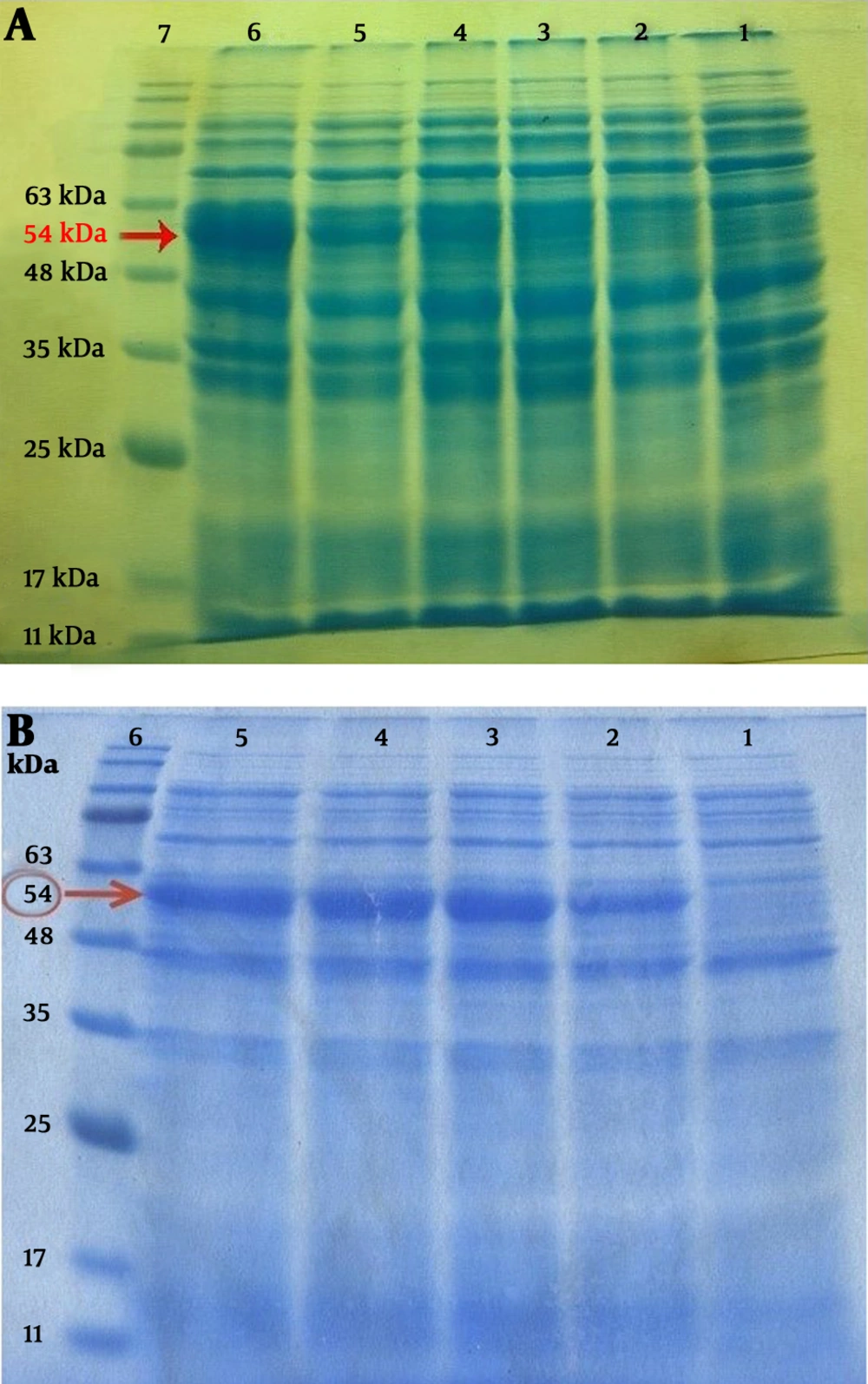
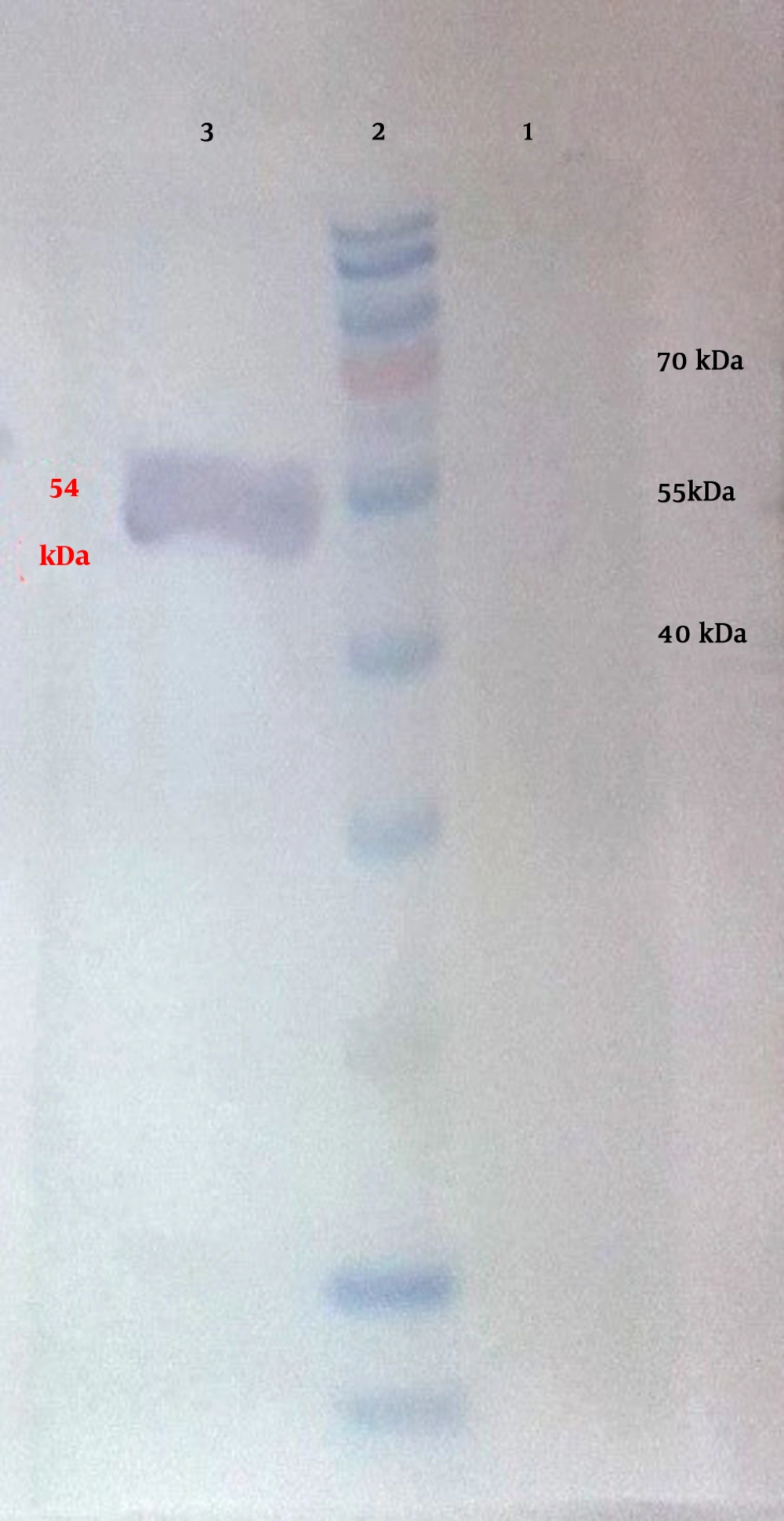
The 3M2e fragment was cut from pET28a+3M2e plasmid with BamHI digestion. After extraction and purification of the 3M2e fragment and pET28a+Flic linearizing by the same enzyme, ligation was performed by T4 ligase to construct the pET28a+3M2e.FliC recombinant plasmid. Following the transformation of this plasmid to ER2566 and BL21 (DE3), colony-PCR was performed to identify the transformants. The correct oriented clones amplify a 634 bp band. The results of SDS-PAGE for the TCP samples after the induction through a time course indicated a gradual increase in the intensity of the 54 kDa band corresponding to 3M2e.FliC fusion protein. Figure 5 shows the results of expression in Bl21 (DE3) and ER2566 strains, respectively. The relative expression levels of 3M2e.FliC 4 h post-induction were estimated based on at least 14.9% and 36% of the total cell protein of Bl21 (DE3) and ER2566 strains, respectively. Western blotting also confirmed the expected polypeptide bands with molecular weight of 54 kDa, which were His-tag fused in N-terminal (Figure 6).
A, 12% SDS-PAGE gel of 3M2e.FliC protein expression in BL21 (DE3) strain. Fractions of total cell protein (TCP) were collected before and after induction through a time course. Lane 1, before isopropyl β-D-1-thiogalactopyranoside (IPTG) induction; lanes 2 to 6, collected samples 1 h, 2 h, 3 h, 4 h and overnight after IPTG induction, respectively. Lane 7, prestained protein ladder (CinnaGen Cat#PR911654). B, 12% SDS-PAGE gel of 3M2e-FliC protein expression in ER2566 strain. Fractions of TCP were collected before and after induction through a time course. Lane 1, before IPTG induction; lanes 2 to 5, collected samples 1 h, 2 h, 3 h and 4 h after IPTG induction, respectively; lane 6, prestained protein ladder (CinnaGen Cat#PR911654).
The result of western blot of 3M2e.FliC protein expressed in ER2566 strain using Anti-His6 monoclonal antibody peroxidase conjugate. Lane 1, total cell protein (TCP) sample of transformed cells before isopropyl β-D-1-thiogalactopyranoside (IPTG) induction (t0); lane 3, TCP sample 4 h after IPTG induction, showing expected 54 kDa 3M2e.FliC fused with His-tag band; lane 2, fermentas prestained protein ladder (#00196905).
5. Discussion
Universal influenza A vaccines that protect against viruses of all subtypes are highly desirable. Antibodies against the highly conserved ectodomain of the M2 ion channel protein (M2e) are broadly cross-reactive (24). Because of the low immunogenicity in nature, M2e is often fused with a larger carrier to enhance anti-M2e immune responses in vaccination experiments. The hepatitis B virus core protein, Neisseria meningitidis outer membrane protein, HSP70 of Mycobacterium tuberculosis, and papaya mosaic virus coat protein (PapMV-CP) as a molecular adjuvant are applied to enhance the immune response to the M2e epitope (25-29). FliC, which in this work was N-terminally coupled with three tandem repeats of the M2e gene, is a molecular adjuvant known as a potent stimulator of the immune system, particularly in the respiratory system (30). The molecular weight predicted for the recombinant protein 3M2.FliC is 54.6 kDa. Because of the isoelectric pH = 5.74, this protein is considered to be almost acidic. The acidity of a protein in the expression system can be considered as an advantage, because in the purification steps and using a pH gradient, this target protein could be isolated more easily from the background proteins.
To design a recombinant peptide vaccine, one of the most important tasks is to ensure that the epitopic regions of the recombinant peptide are located on or near the protein surface. Our results clearly indicated that most regions of 3M2e sequence (1 - 120 residue numbers) and the partial region of FliC were realized as linear epitopes in the context of a B cell response. In agreement with the results of the linear epitope prediction, the superficial amino acids prediction of 3M2e.FliC fusion antigen demonstrated that the recognized linear epitopes were accessible at protein surface areas, a desired characteristic of an antigen to elicit immune responses efficiently. In this study, the valid servers such as I-TASSER and Phyre2 and Modeller v9.15 software were used for protein modeling construction. The validity of these models was assessed using the Q-mean server. Therefore, the final models presented here, which gained higher scores than the other ones, are the best-constructed models. Ramachandran plot analysis also confirmed their good quality. Meanwhile, these models show that the 3M2e epitopes are available on the surface of protein with convenient positions available for being recognized by the immune system.
The presence of a poly-histidine tag upstream of the T7 promoter in the pET28a vector led this tag to be added to the N-terminal of 3M2e.FliC. This fusion allowed us to detect the 3M2e.FliC recombinant protein in western blot and to purify it by Ni-NTA resins in follow-on stages. Based on our cloning strategy, a thrombin protease site has been located downstream of the poly-histidine tag, which allows to remove poly-histidine tag from 3M2e.FliC through enzymatic digestion. It has been reported that the ER2566 strain of E. coli has less background expression in comparison with Bl21 (DE3) (31). Our results showed more yield of 3M2e.FliC with less background proteins when the clone was expressed into ER2566 strain. A recent study has reported the expression of 3M2e.FliC in Bacillus subtilis as a secretion system which may shortcut the purification processes of 3M2e.FliC (32). Follow-on studies are needed to optimize the expression conditions, purify this fusion protein, and assess it as an influenza vaccine candidate in animal models alongside proper control groups.
5.1. Conclusions
In silico analysis showed plenty of surface-exposed epitopes on the 3M2e.FliC protein, a desired structure capable of inducing immune responses efficiently. This recombinant antigen may be considered as a universal influenza vaccine candidate after its evaluation and assessment in animal models. Expression experiments also indicated that ER2566 strain was more efficient for the production of the 3M2e.FliC antigen when compared to the BL21 (DE3) strain.