1. Background
It is estimated that 36 million people will die due tuberculosis disease by the year 2020 (1). Mycobacterium bovis Bacillus Calmette-Guerin (BCG), the only licensed vaccine that has been used globally for protection of children, in disseminated tuberculosis form, for more than 80 years, has variable efficacy in adults with pulmonary tuberculosis (2). One-third of the world’s population has latent tuberculosis infection and is at risk of developing active tuberculosis (3). Most often, the strategy used in the design of vaccine candidates in clinical trials to prevent active tuberculosis is prophylaxis, but postexposure vaccine strategy is based on latent tuberculosis infection to prevent the reactivation of dormancy phase of the disease (4).
DosR regulon is a set of nearly 50 genes introduced in 2003, and its high expression is dependent on hypoxia and nitric oxide in the environmental conditions that inhibit the growth of M. tuberculosis (5, 6). Some studies have shown that DosR regulon antigens are upregulated during the persistence phase of the disease in the mouse model (6), and people with latent tuberculosis infection have DosR regulon antigen specific T- cells compared to those with active infection (7). Therefore, Regulon DosR antigens could be suitable candidates for developing postexposure vaccines against tuberculosis.
Because Th1 cell mediated immune response plays an important role against tuberculosis infection (8), many experimental and bioinformatics analyses were performed to identify strong T- cell mediated immunogenic antigen. Previous studies have revealed that Rv2029c, Rv2031c, and Rv2627c antigens of Regulon DosR have the potential to induce strong T- cell immune response (9). Rv2029c antigen or the pfkB gene, which is considered to produce phosphofructokinase B, was shown to significantly induce strong T- cell response in tuberculin skin test positive individuals (10).
Rv2031c or the hspX gene (α-crystallin or acr) is an immunodominat antigen among the proteins expressed during the latency phase of tuberculosis. The efficacy of this protein as a qualified vaccine candidate was shown in various studies (11, 12). Rv2627c is a conserved hypothetical protein that induces Th1 immune response and is significantly recognized in tuberculin skin test positive individuals (10). In addition, the basic purpose of tuberculosis vaccine development is to induce immune response by production of long-lived memory T- cells. Lack of long-lasting T- cells is the main reason for BCG not to provide protection for the individuals with latent tuberculosis infection (13). It has been shown that among regulon DosR antigens, Rv2031c and Rv2627c latency antigens are prominent players in inducing this population of long-lasting central memory T- cells (14).
Microtubule-associated protein light chain 3, also called autophagy-related gene 8 (Atg8), is one of the major components of the autophagy system involved in macroautophagy activity (15). New studies indicate that autophagy significantly increases the processing and presenting antigens by MHC class II molecules (16, 17). Thus, fusion of targeting antigens with microtubule-associated protein light chain 3 may increase CD4 T- cell response in vaccine design.
DNA vaccine is a new technology in which foreign DNA is transported to the nucleus of Myocytes and antigen presenting cells (APCs) by the intramuscular route (18). DNA vaccine strategy triggers both CD4 and CD8 T- cell immunity response to provide optimized vaccine response compared to old conventional vaccine strategies (19). DNA vaccination strategy has been used as a valuable vehicle for evaluation of MHC class I-and class II-restricted epitope of mycobacterial antigens (20). In this regard, bioinformatics tools provide epitope prediction and development of epitope-driven DNA vaccines that proved successful in various group studies (19).
In the current study, bioinformatics tools were used to design a multi-epitope DNA vaccine based on MHC class I-and class II-restricted T- cell epitopes of 3 latency associated antigens (Rv2029c, Rv2031c, and Rv2627c) combined with microtubule-associated protein light chain 3. In addition, we analyzed peptide expression by bioinformatics tools, and our in silico results indicated efficacy of this new postexposure vaccine.
2. Methods
2.1. Primary Sequence and Homology
Primary sequence of the 3 latency-associated antigens (Rv2029c, Rv2031c, and Rv2627c) and microtubule-associated protein light chain 3 were obtained from NCBI (www.ncbi.nlm.nih.gov) and used for in silico analysis. The physic-chemical characteristics of each protein were retrieved from Uniprot (www.uniprot.org), and protparam (us. expasy.org/tools/ protparam.html). BLAST (BLAST.ncbi.nlm.nih.gov) was performed for the 3 latency-associated antigen sequences against nonredundant proteins.
2.2. Immunoinformatics Analysis
2.2.1. Prediction of MHC Class I-Binding Peptides
There are 8 different methods for predicting MHC class I-binding peptides, which are as follow: Artificial neural network (ANN); average relative binding (ARB); stabilized matrix method (SMM); stabilized matrix method with a peptide: MHC binding energy covariance matrix (SMMPMBEC); scoring matrices derived from combinatorial peptide libraries (Comblib-Sidney2008); Consensus; and NetMHCpan and IEDB. We used 3 online tools to identify MHC class I epitopes which applied more methods. IEDB at tools.immuneepitope.org/analyze/html/mhc_binding.html is a server that contains characteristics of all methods. RANKPEP at imed.med.ucm.es/Tools/rankpep predicts MHC class I-binding peptides by implying position- specific scoring matrices (PSSMs); and nHLAPred is a comprehensive tool to predict MHC class I-binding peptides, which implies artificial neural networks (ANNs) and quantitative matrices (QM) options; QM is a combination of artificial neural networks and quantitative matrices. The cut-off value for IEDB based on the percentile rank method was considered as < 1%, ≥ 16 for RANKPEP and ≥ 16 for nHLApred.
2.2.2. Prediction of MHC Class II-Binding Peptides
MHC2Pred at www.imtech.res.in/raghava/mhc2pred/ is a support vector machine (SVM) prediction method that identifies promiscuous MHC class II-binding peptides. This server along with RANKPEP at http://imed.med.ucm.es/Tools/rankpep was employed to identify MHC class II-binding peptides. The cu-off scores were chosen as ≥ 16 for RANKPEP, and ≥ 0.5 for MHC2Pred.
2.2.3. Analyzing Epitopes for Human HLAs Binding and Population Coverage
MHC class I-and class II-binding epitopes analyzed human HLAs binding. Screening was performed by IEDB server. The SMM and ANN were applied, with a cut off of IC50<500nM. It has been shown that an IC50 < 500nM value assents more than 80% accuracy in MHC peptide binding prediction (21). Population coverage was also calculated by IEDB server.
2.3. Peptide Construct Design and Analysis Associated Feature
2.3.1. Primary Peptide Construct Design
Mouse construct was designed based on MHC class I-and class II-restricted epitopes of the 3 latency-associated antigens of Rv2029c, Rv2031c, and Rv2627. MHC restricted epitope segments were linked together by appropriate linkers.
2.3.2. Sequence-Based Primary Structure Analysis
Wide range of primary characteristics of peptide construct was analyzed by various tools. ProtParam at web.expasy.org/protparam/ was used to identify the physic-chemical parameters of a protein, and ePEST at emboss.bioinformatics.nl/cgi-bin/emboss/epestfind was used to identify PEST sequence.
2.3.3. Secondary-Structure Analysis
Predict Protein server at www.predictprotein.org/ is the server used for secondary structure prediction. Different features of secondary structure such as coiled coils and zippers were analyzed. ExPASy COILS server at embnet.vital it.ch/software/COILS_form.html and Paircoil2 at groups.csail.mit.edu/cb/paircoil2/paircoil2.html were used to predict coiled coils. ExPASy COILS works based on matching query sequence with known standard coiled coils, measuring a similarity score (22). Paircoil2 identifies coiled coil motifs by using potential pairwise residue (23). 2ZIP at 2zip.molgen.mpg.de/ is a prediction server for zippers that incorporates a standard prediction algorithm by searching for the existence of Leucine repeats (24).
2.3.4. Tertiary Structure Prediction
Prediction of 3D structure was performed by I-TASSER server at zhanglab.ccmb.med.umich.edu/I-TASSER/.I-TASSER. I-TASSRR performs structure prediction in 3 stages starting as multiple threading alignments, iterative structural assembly, and finally protein function obtained from structural-structural matching of the target by known proteins. Two specific parts of the tertiary structure are domains and motifs. Domains fold independently and have particular functional roles, and motifs are specific part of the protein that do not fold independently. InterProScan at www.ebi.ac.uk/Tools/pfa/iprscan/ was launched to identify domains/motifs (25).
2.3.5. Homology Analyses
Peptide construct was BLAST against nonredundant proteins in both protein sequence and PDB database of NCBI.
2.3.6. Posttranslational Modification, Protein Sorting and Localization
Several servers were utilized for the prediction of specific posttranslational modification (PTMS). Prediction of different types of modifications (Table 1) such as lipid PTMs, phosphorylation, and glycosylation were performed on the peptide construct sequence. Moreover, proteins were sorted to different subcellular parts in cotranslational or posttranslational phases. Some of the tools were used to predict protein sorting and localization as well as PTMs (Table 1).
| Specific PTMs | |||
|---|---|---|---|
| Specific function predictor | Server | URL | |
| Lipid PTMs | GPI lipid anchoring | big-PI/GPI animals (19 - 28) | mendel.imp.ac.at/gpi/gpi_server. html |
| Myristoyl | MyrPS/NMT (20 - 28) | mendel.imp.ac.at/myristate/SUPLpredictor.html | |
| Prenyl anchors | PrePS (36-28) | mendel.imp.ac.at/sat/PrePS | |
| Phosphorylation | General phosphorylation site | NetPhos (57) | www.cbs.dtu.dk/services/NetPhos |
| Kinase-specific phosphorylation sites | NetPhosK (56) | www.cbs.dtu.dk/services/NetPhosK | |
| Glycosylation | N-glycosylation sites | NetNGlyc (64) | www.cbs.dtu.dk/services/NetNGlyc |
| Mucin-type GalNAc O-glycosylation sites | NetOGlyc (65) | www.cbs.dtu.dk/services/NetNGlyc | |
| C-mannosylation sites | NetCGlyc (69) | www.cbs.dtu.dk/services/NetCGlyc | |
| Epsilon amino residue Lysine Glycation of epsilon amino groups of lysines in mammalia | NetGlycate (70) | www.cbs.dtu.dk/services/netGlycate | |
| OβGlcNAc attachment sites | YinOYang (64) | www.cbs.dtu.dk/services/YinOYang | |
| Protein Sorting | |||
| Signal peptide | Signal peptide and cleavage sites | SignalP | www.cbs.dtu.dk/services/SignalP |
| Protein subcellular localization | Subcellular localization of proteins | PSORT | psort.hgc.jp/ |
| Subcellular localization of proteins in eukaryotes | CELLO | cello.life.nctu.edu.tw/ | |
| Subcellular localization of proteins in eukaryotes | BaCelLo | gpcr.biocomp.unibo.it/bacello/ | |
2.3.7. Allergenicity Properties
The IgE epitopes and allergic properties of the mouse construct were analyzed by AlgPred at www.imtech.res.in/raghava/algpred/index.html (26) and PREAL at gmobl.sjtu.edu.cn/ PREAL/index.php (27). AlgPred is a SVM- based server with large amounts of allergens and nonallergens database that predict allergen property of a protein with high specificity according to existence of IgE epitopes. PREAL is another SVM- based allergen predictor server that determines potential allergenicity based on key factors. The server implies various physic-biochemical characteristics to achieve a high accuracy in prediction, ranging from 93.42% to 100%.
2.3.8. Antigenicity Characterizatio
ANTIGENpro at scratch.proteomics.ics.uci.edu/ was used as a first sequence-based and only alignment-free predictor that used large amounts of nonredundant data obtained from analyzing protein microarrays (28). Another server was also used: VaxiJen at www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html. Furthermore, VaxiJen was used as the first alignment-independent predictor server for protective antigens based on physic-chemical properties of a protein (29).
2.3.9. Development of Final Peptide Construct
To design the final peptide construct as a vaccine candidate, microtubule-associated protein light chain 3 was added to the primary peptide construct. All analyses in the present study were performed for both the primary and final construct to identify the effect of microtubule-associated protein light chain 3 in addition to the primary construct.
2.4. Gene Construct
2.4.1. Reverse Translation and Codon Optimization
JCAT at www.jcat.de (30) is a server with the simultaneous possibility of reverse translation and codon optimization.
2.4.2. mRNA Structure Prediction
Vfold server at rna.physics.missouri.edu provides the possibility to predict RNA structure, thermodynamics- based folding and stability (31).
3. Results
3.1. Sequence and Homology Analysis
Sequence of the 3 latency-associated antigens of Rv2029c, Rv2031c, and Rv2627c and mouse microtubule-associated protein light chain 3, respectively, with accession numbers of P9WID3.1, P9WMK1.1, P9WL67.1, and Q9CQV6.3 were retrieved from NCBI. Physicochemical properties of each protein were obtained from Uniprot and protparam, and all features for each protein were desirable as a candidate protein. BLAST performed for the 3 latency-associated antigens showed a more than 99% homology between M. tuberculosis complexes for each protein.
3.2. Immunoinformatics Assay
3.2.1. Identifying MHC Class I-and Class II-Restricted T- Cell Epitopes
Epitopes were predicted over multiple alleles of Balb/c H-2 Class I as H2-Kd, H2-Dd and H2-Ld. The epitopes were selected based on high score and overlapped between the 3 online software predictors (Table 2). In total, 5 regions were selected which were located in Rv2029c (44 - 57 and 115 - 127), Rv2031c (60 - 68 and 96 - 104), and Rv2627c (27-35). Three- associated latency antigens were also analyzed to predict the MHC class II epitopes. Epitopes were predicted over Balb/c H-2 Class II alleles as H2-IAd and H2-IEd. Five regions were selected based on the high score and overlapped between the 3 software predictors (Table 3). Regions were placed in Rv2029c (135 - 149 and 253 - 264), Rv2031c (118 - 132), and Rv2627c (325 - 339 and 390 - 404).
| Protein | Selected Epitope Region | MHC Class I Epitope Binding Prediction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IEDB | nHLApred | RANKPEP | ||||||||
| Position | Allele | Score | Position | Allele | Score | Position | Allele | Score | ||
| Rv2029c | RYDPGGGGINVARI | 49 - 57 | H2-Dd | 0.8 | 49 - 57 | H2-Dd | 23.690 | 44 - 52 | H2-Kd | 20.705 |
| RFVLPGPSLTVAE | 119 - 127 | H2-Dd | 0.4 | 119 - 127 | H2-Dd | 23.530 | 116 - 126 | H2-Kd | 21.269 | |
| Rv2031c | VDPDKDVDI | 60 - 68a | H2-Dd | 1.4 | 60 - 68a | H2-Dd | 13.110 | 60 - 69 | H2-Dd | 19.155 |
| GSFVRTVSL | - | - | - | 94 - 104 | H2-Dd | 16.91 | 94 - 104a | H2-Dd | 13.646 | |
| Rv2627c | GPFMHTGLY | 27 - 34 | H2-Dd | 0.2 | 26 - 34 | H2-Dd | 18.110 | - | - | - |
aPredicted epitope near the cut-off to show overlap with the selected epitope which predict by the other tool.
| Protein | Selected Epitope Region | MHC Class II Epitope Binding Prediction | |||||
|---|---|---|---|---|---|---|---|
| MHC2Pred | RANKPEP | ||||||
| Position | Allele | Score | Position | Allele | Score | ||
| Rv2029c | LRGAAASAAFVVASG | 141 - 149 | H2-IAd | 0.683 | 138-146 | H2_IAd | 16.131 |
| IPMTAVSGVGAG | 256 - 264 | H2-IAd | 0.684 | - | - | - | |
| Rv2031c | DKGILTVSVAVSEGK | - | - | - | 118 - 126 | H2-IAd | 16.713 |
| Rv2627c | IGRMISPLSLTPLVP | 324 - 332 | H2-IAd | 0.545 | - | - | - |
| RFVQAALEQSGLLDA | 393 - 404 | H2-IAd | 0.575 | 390 - 398a | H2-IAd | 12.638 | |
aPredicted epitope near the cut-off to show overlap with the selected epitope which predict by the other tool.
3.2.2. Human HLAs Binding Alleles and Population Coverage Analysis
The number of human HLAs binding alleles and population coverage for each predicted epitope were retrieved from IEDB server (Table 4). Predicted epitopes showed high promiscuity in considering the number of human HLAs binding alleles. Population coverage for all the epitopes was calculated globally, and the results revealed a broad spectrum with more than the average of 50% frequency (Table 4).
| Protein | Epitope | No. of HLA Binding Alleles | Population Coverage Calculation Result | ||
|---|---|---|---|---|---|
| Coverage, %a | Average hitbb | PC90c | |||
| RV2029c | RYDPGGGGINVARI | 14 | 67.94 | 0.94 | 0.31 |
| RFVLPGPSLTVAE | 18 | 51.52 | 0.64 | 0.21 | |
| LRGAAASAAFVVASG | 12 | 34.03 | 0.36 | 0.15 | |
| IPMTAVSGVGAG | 27 | 53.05 | 0.61 | 0.21 | |
| Rv2031c | VDPDKDVDI | 9 | 21.35 | 0.22 | 0.13 |
| GSFVRTVSL | 18 | 74.94 | 1.09 | 0.40 | |
| DKGILTVSVAVSEGK | 8 | 45.04 | 0.50 | 0.18 | |
| RV2627c | GPFMHTGLY | 21 | 56.85 | 0.74 | 0.23 |
| IGRMISPLSLTPLVP | 10 | 65.37 | 0.79 | 0.29 | |
| RFVQAALEQSGLLDA | 9 | 33.83 | 0.36 | 0.15 | |
aProjected population coverage.
bAverage number of epitope hits / HLA combinations recognized by the population.
cMinimum number of epitope hits / HLA combinations recognized by 90% of the population.
3.3. Peptide Construct Development and Analysis
3.3.1. Primary and Final Peptide Construct Development
Primary mouse construct was designed based on 2 segments: MHC class I-restricted epitopes and MHC class II-restricted epitopes. MHC class I-restricted epitopes fused together tandemly by AAY linkers and were placed at the N-terminus of the construct. Also, MHC class II-restricted epitopes were linked by GPGPG linkers and located at the C-terminus. Final construct was developed by the direct fusion of C-terminus of primary construct with N-terminus of LC3 (Figure 1).
Structure of Amino Acid Sequence of Final Construct. MHC Class I and II Epitopes of Antigens Which Joined Together by Appropriate Linkers Indicating the Position of Epitopes. AAY Linkers Was Applied to Fuse MHC Class I-Restricted Epitopes, GPGPG Linkers Was Utilized to Link MHC Class II-Restricted Epitopes and LC3 Directly Fused to MHC Class II-Restricted Epitopes. Linkers Showed With Highlight and LC3 Showed with Underlined Sequence
3.3.2. Primary Sequence Analysis
Basic physicochemical characteristics of the primary and final constructs were analyzed by the protparam server (Table 5). There were no significant PEST regions in the primary or in the final construct.
aTotal number of negatively charged residues (Asp + Glu).
bTotal number of positively charged residues (Arg + Lys).
cMammalian reticulocytes, in vitro.
dGrand average of hydropathicity.
3.3.3. Secondary and Tertiary Structure Prediction
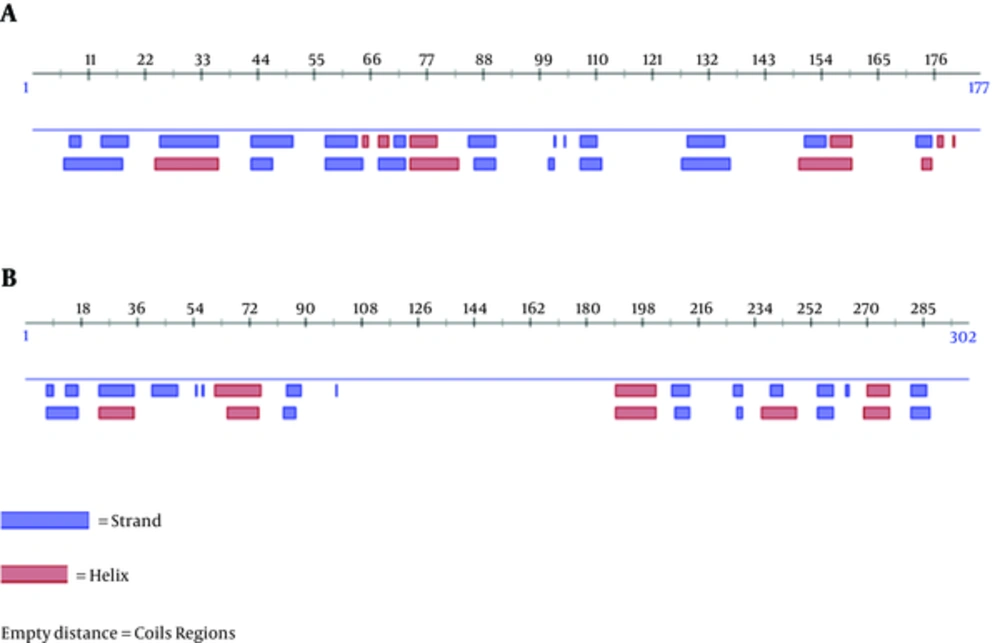
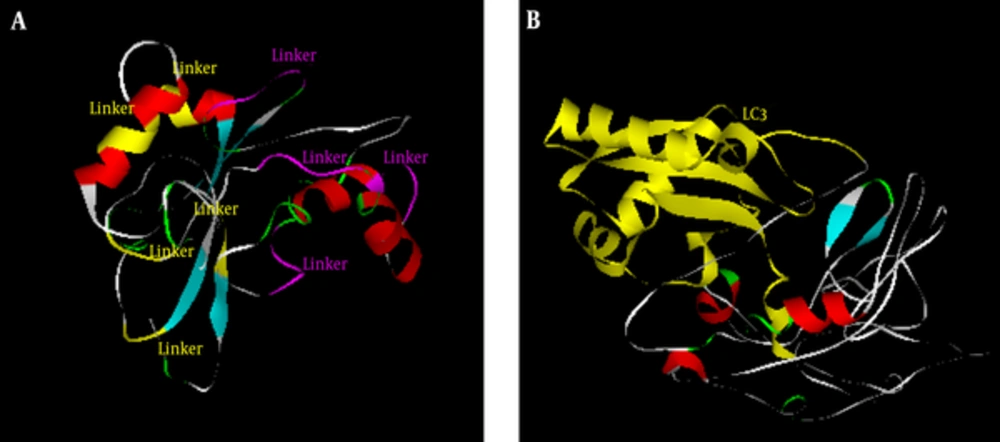
The secondary structure of the primary and final constructs was predicted by Predict Protein (Figure 2). The results revealed that the secondary structure of the primary construct consisted of 20.9% helix, 29.9% strand, and 49.2% loop, and the final construct consisted of 19.2% helix, 12.6% strand, and 68.2% loop. There were no no coiled coil and Leucine Zipper domain in the primary or in the final constructs based on coils, Paircoil2, and 2ZIP servers. The tertiary structure of the primary and final constructs was modeled by I-TASSER server (Figure 3). Five models were predicted for the primary and final constructs; and for both constructs, Mode l was considered as the best model based on C-score (-2.51 for primary and -3.49 for final construct). Moreover, the analysis of the primary and final construct sequences by InterProScan showed no protein domain in the primary structure, and there was only one domain in the final construct as ubiquitin-related domain that associated to LC3.
3.3.4. Homology Analyses
The primary peptide construct was BLASTed against nonredundant protein sequences in NCBI. The primary construct BLAST determined no putative domain and revealed more than 60% homology with phosphofructokinase of M. tuberculosis. The final construct was BLASTed against PDB databases in NCBI, and the results showed only homology with ubiquitin-related domain.
3.3.5. Posttranslational Modification, Protein Sorting and Localization Prediction
A wide range of modification predictions was analyzed in both primary and final constructs. There was no lipid PTMs as GPI-modification, N-terminal glycines myristoyl, or prenylation in the primary and final constructs. Phosphorylation modification analysis showed 8 and 14 modification sites in the primary and final constructs, respectively. Various glycosylation modifications were determined. NetGlycate predicted 1 glycosylation in position 169 in the primary construct, and 4 glycosylation in positions 169, 185, 207, and 242 in the final construct. NetOGlyc predicted 1 glycosylation in position 167 in the primary construct, and 3 in positions 128, 167 and 180 of the final construct. YinOYang predicted O-GlcNAc sites in 6 and 7 positions of the primary and final constructs, respectively. According to the results of SignalP, there was no signal peptide in the primary and final constructs. Subcellular localization prediction by several servers revealed that both primary and final peptide constructs had cytoplasmic localization with high reliability scores.
3.3.6. Allergenicity and Antigenicity Characterization
Allergenicity of the primary and final constructs was evaluated and the results indicated that both constructs were nonallergen with negative predictive value of 94.18%. The primary and final constructs antigenicity was estimated by ANTIGENpro (0.720 and 0.228) and by VaxiJen (0.7475 and 0.5418).
3.4. Gene Construct Development and Analysis Associated Features
The final peptide construct was utilized to develop gene construct. JCAT server was used for a simultaneous reverse translation and codon optimization. Gene construct consists of 858 nucleotides optimized based on the mouse genetic code with 69.34% GC content and 0.71 CAI-Value. Additional elements such as kozak sequence, restriction sites, start, and stop codons were added to the gene construct; mRNA stability analysis was done on the optimized mouse construct. Thermodynamic prediction showed -439.46 kcal/mol. Moreover, the 10 nucleotides at the start of the 5’end were not in the secondary structure with ∆G < -10.
4. Discussion
Most current vaccine candidates evaluated in clinical trials are prophylactic vaccines, which work when administered in uninfected individuals to prevent the active form of tuberculosis. According to the WHO report in 2015, it is estimated that one-third of the world population was latently infected with M. tuberculosis. Therefore, the postexposure vaccine strategy, which focuses on latent tuberculosis infection to prevent progression to active disease, is necessary (4). DosR regulon antigens were shown to qualify as candidates to be utilized in postexposure vaccine strategy designs (4). Three latency-associated antigens (Rv2029c, Rv2031c, and Rv2627c) are common in all studies as strong T- cell antigens; therefore, we selected them in our study. In addition to the chimeric protein technology, production of multi-epitope vaccines has similar beneficial advantages. In multi-epitope based vaccines, immunodominat epitopes are included in the vaccine to enhance the efficacy of the vaccines (32).
Because immune protection against tuberculosis is based on Th1 cell-mediated immune responses by CD4 and CD8 T lymphocytes, we used immunoinformatics tools to determine MHC class I-and class II-restricted binding epitopes. Furthermore, microtubule associate light chain 3 was implied to enhance the presentation of antigens by MHC class II molecules to CD4 T- cells, which are the main T- cell types in protection against tuberculosis (33).
The main challenge to develop multi-epitope based vaccines is applying bioinformatics to identify immunoprotective epitopes. Because the attachment of T- cell epitopes to MHCs is in a linear form, this interaction can be modeled successfully with high accuracy (34). Based on this interaction, a large number of algorithms were developed for T- cell epitope mapping (35). Various MHC class I-restricted T- cell epitope predictor tools have been developed that cover large numbers of alleles and have high accuracy prediction value, ranging from 90% to 95% (36). Among the large number of predictor servers for MHC class I-restricted epitopes, RANKPEP, IEDB, and nHLAPred were selected in this study.
Although there are large numbers of servers that predict MHC class I epitopes with high accuracy, only a few servers completely achieve MHC class II epitopes (37). There are 3 servers (MHC2Pred, RANKPEP and IEDB) that most comprehensively predict MHC class II-binding epitopes (38). The high score epitopes that were common between the servers were selected. In addition, we used the IEDB server to identify the number of human HLAs that potentially bind to predicted epitopes and estimate the population coverage for each epitope. In this line, the predicted epitopes had the potential of binding to large number of human HLAs, with a broad range of population coverage.
Finally, the primary and final constructs were designed. Protparam analysis showed high aliphatic index in both primary and final constructs, indicating thermo stability. Primary construct has low instability index and is classified as a stable protein, but the final construct with high instability index is classified as unstable protein due to instability of LC3. Thus, LC3 preserves instability features in the final construct, leading to the degradation of the protein and subsequent entrance into the MHC pathway (39, 40). ProteinPredict server analysis revealed that the secondary structure of the primary and final constructs contained helix, strand, and loop, and the final construct had more loops compared to the primary construct. Loop structures are involved in various biological functions. Linkers and LC3 segments have functions associated with loops in the constructs. The LC3 structure consists of large numbers of loops that is important for LC3 processing in autophagy, so the existence of this segment in the final construct resulted in high a number of loops (41). Tertiary structure of the constructs were modeled by I-TASSER and based on z-score. The final construct is believed to correlate with the conformation of LC3 crystal structure (42).
Posttranslational modifications (PTMs) are various numbers of chemical changes that modify structures. Charge and conformation of a protein can lead to a change in binding affinity, enzyme activity, and hydrophobicity (43). In this regard, we analyzed both constructs for 3 categories of PTMs. In lipid PTMS, GPI-modification, N-terminal glycines myristoyl, and prenylation were analyzed. GPI anchored modification is related to altered antigenicity binding and protein interactions with membrane (44). N-terminal glycines myristoyl and prenylation are acylation modification of a protein, and these groups of modifications result in protein hydrophobicity change and target the protein to the surface of cell membrane (45). The results of the analysis revealed that neither the primary nor the final constructs undergo lipid modifications.
Phosphorylation modifications are considered as main modulators of signal transduction and are associated with various types of protein function in cellular networks such as regulating cellular metabolism, survival, apoptosis, and enzyme activity (46). Several phosphorylation modifications have been shown to be launched by eukaryotic cells to regulate ATG8/LC3 family of proteins (47). There were several phosphorylation modifications in the primary and the final constructs which indicate preservation of LC3 function in conjugate form in the construct. In fact, existence of phosphorylation sites lead to better degradation and final epitope presentation (48). Different servers analyzed various glycosylation modifications, and several glycosylation modifications were predicted for both the primary and final constructs. The number of O-and N-linked glycosylation in the final construct is due to the role of these types of modifications in autophagy regulation of the ATG8/LC3 family. Moreover, according to experimental studies, glycosylation modifications have no role in antigenicity and potency of a DNA vaccine (49).
The signal peptide is a short sequence of amino acids at the N-terminus of proteins and leads to secretory pathways (50). Analysis of primary and final constructs did not show any peptide signal on either constructs to enable them to reside inside organelles such as Golgi or Endoplasmic reticulum. Subcellular localization prediction revealed that both primary and final constructs have cytoplasmic localization with high reliability score. This type of localization results in immune induction by interaction with the MHC class I and class II pathways (51).
The final construct was utilized to perform codon optimization based on the mouse genetic code for optimal expression (52). One of the key points in gene expression is mRNA stability with more stability in mRNA, resulting in more protein expression. The parameter that indicates stability is measure of ∆G, with a lower index leading to higher stability (53). Because mRNA from a final mouse construct had low ∆G, the stability of the mRNA was confirmed. Moreover, 10 nucleotides at the mRNA 5’end are not in secondary structure, and this is the reason for ∆G < -10, which is important in predicting the initiation of translation, thus, translation will be performed with efficiency (54).
A wide range of in silico analyses were performed on the primary and final constructs, indicating that the addition of LC3 with the function of presentation epitopes to MHC class II had no adverse effect on antigenicity, structural stability, signal peptide, and subcellular localization. Thus, the final construct is introduced as a qualified postexposure vaccine candidate, with improvement in presentation of epitopes to the immune system. Bioinformatics analyses indicated the designed vaccine has strong potential to be evaluated for immunogenicity and protective efficacy in the experimental model to introduce novel postexposure vaccine candidate for people with latent tuberculosis infection.