1. Background
Streptococcus pneumoniae causes a wide range of infectious diseases, including community- and hospital-acquired pneumonia, which is globally associated with high morbidity and mortality rates (1-3). It has been found that S. pneumoniae can adhere to lung cells following the formation of aggregates on the cell surface via many adhesins, including pneumococcal serine-rich repeat protein (PsrP), a large surface glycoprotein first identified in S. pneumoniae serotype 4, isolate TIGR4 (2, 3). Pneumococcal serine-rich repeat protein contains five important domains including two serine-rich repeat motifs of SAS [A/E/V] SAS [T/I] (2, 3).
Pneumococcal serine-rich repeat protein homologs include serine-rich adhesin for platelets (SraP) and fimbriae-associated protein 1 (Fap1), which are located on the cell surface of Staphylococcus aureus and S. parasanguinis FW213, respectively; they have also been demonstrated to be glycoproteins (4-8). In S. aureus, a SraP-SecY2A2 island including SraP and seven other genes located downsstream of the SraP was shown to be required for SraP glycosylation and secretion, and glycoslytransferase A (GtfA) and glycosyltransferase B (GtfB) have been shown to be involved in the glycosylation of recombinant SraP (5). In S. parasanguinis FW213, Fap1 and 11 other genes construct a Fap1-SecY2A2 island to regulate the glycosylation and secretion of Fap1, and a Gtf1-Gtf2 complex initiates Fap1 glycosylation (7-9). In S. pneumoniae TIGR4, PsrP and 17 other genes located downstream of PsrP construct a PsrP-SecY2A2 island, and GtfA and GtfB have been found to initiate PsrP glycosylation (2, 10).
Glycosyltransferase A has been demonstrated to be an O-GlcNAc transferase involved in PrsP glycosylation, whereas GtfB may function as a co-activator for the full activity of GtfA (2, 10). An in vitro hydrolytic assay showed that the activity of the mixture of GtfA and GtfB was 100-fold higher than that of the individual GtfA, but GtfB had no any activity (10). Mutations of GtfA, including N98A, E244A, E332A and S403A, block the glycosylation of PsrP (2, 10). To date, 94 S. pneumoniae serotypes have been found (11-14). Previous studies showed that PsrP was only detected in 51.7% of invasive S. pneumoniae isolates and in 48.3% of carrier S. pneumoniae isolates (15), thus, we sought to examine if the presence of the PsrP-SecY2A2 island is associated to a certain serotype of S. pneumoniae, or if GtfA and GtfB are conserved among these S. pneumoniae isolates and involved in the glycosylation of PsrP via the formation of the glycosyltransferase complex GtfA-GtfB.
2. Objectives
In the present study, we aimed to investigate the distribution of serotypes and the prevalence of the PsrP-SecY2A2 island in S. pneumonias isolates, estimate the evolutionary divergence of GtfA and GtfB among different isolates using the MEGA4 software package, construct an in vivo glycosylation system in Escherichia coli to identify whether GtfA and GtfB contribute to the glycosylation of PsrP, and investigate the interaction of GtfA with GtfB using a glutathione S-transferase (GST) pull-down assay and yeast two-hybrid experiment.
3. Methods
3.1. Ethics Statement
The Ethics Committee of Zhongnan Hospital, Wuhan University, approved this study (2017S011).
3.2. Sample Collection
In total, 141 non-duplicate S. pneumoniae isolates were collected between June 1, 2015 and March 31, 2016. The isolates were derived from sputum (n = 123), blood (n = 12), pharynx swabs (n = 4), and bronchoalveolar lavage fluid (n = 2). All of the isolates were stored in GermBank tubes (Creative Microbiologicals, Ltd., Taipei) at -70°C.
3.3. Streptococcus pneumonia Identification
All S. pneumoniae isolates were identified by microbiological methods, including typical colony morphology, alpha-hemolysis, negative catalase reaction, Gram-positive staining, optochin disk sensitivity, and bile solubility.
3.4. Molecular Methods
3.4.1. Primers and Other Important Reagents
The primers for serotyping S. pneumoniae were designed as previously described (16). Other primers were designed using Primer 3.0 Plus, then further identified by Blast software (Appendix 1 in Supplementary File). Polyclonal anti-S antibody, monoclonal anti-His, and anti-c-Myc antibodies were purchased from MBL (MBL, Japan), Abcam (Abcam, UK), and Santa Cruz Biotechnology (Santa Cruz, USA), respectively. Succinyl wheat germ agglutinin (sWGA) and four other agglutinins labeled with horseradish peroxidase were purchased from EY Laboratories Inc. (EY Laboratories Inc., USA).
3.4.2. Streptococcus pneumoniae Chromosomal DNA Isolation
A Tianamp Bacterial DNA Kit (Tiangen Biotech Co., China) was used to isolate S. pneumoniae chromosomal DNA from the collected isolates following the manufacturer’s instructions. The isolated chromosomal DNA was dissolved in double distilled water (ddH2O) and stored at -70°C for subsequent experiments.
3.4.3. Serotyping of S. pneumoniae
Serotypes 1, 3, 6A/B, 9V, 10A, 11A, 14, 15A, 15B/C, 19A, 19F, and 23F were identified using polymerase chain reaction (PCR) assays and a Bio-rad T100 thermocycler (Bio-rad Laboratories, USA). The final volume of the PCR reaction was 20.0 μL, comprising 2× Taq PCR MasterMix (Tiangen Biotech Co., China; 10.0 μL), a couple of primers at a concentration of 10 μM (1.0 μL), S. pneumoniae chromosomal DNA (1.0 μL), and 8.0 μL of ddH2O. PCR assays were carried out as described in Supplementary File (Appendix 2).
3.4.4. Detection of PsrP, GtfA and GtfB
Pneumococcal serine-rich repeat protein, GtfA, and GtfB were tested using real-time SYBR Green PCR assays and an MXP3000® system (Stratagene, USA). The real-time PCR reaction mixture consisted of 2 × SYBR Premix Ex Taq™ (TaKaRa, China) (10 μL), ROX (0.4 μL), a couple of primers (10 μM, 1.0 μL), S. pneumoniae chromosomal DNA (1.0 μL), and ddH2O (7.6 μL). The optimized amplification conditions are listed in Supplementary File (Appendix 2). Fluorescence was monitored at regular intervals before the denaturation phase. Samples with a cycle threshold value of ≤ 30 were considered to be negative.
3.4.5. Sequencing of GtfA and GtfB
To investigate GtfA and GtfB variations of S. pneumoniae isolates with different serotypes, GtfAB was amplified with an MXP3000® system (Stratagene, USA). The amplification mixture comprised 25.0 μL of 2 × PCR Master Mix (Fermentas, China), 1.0 μL of a couple of primers (10 μM), 2.0 μL of S. pneumoniae chromosomal DNA template, and 22.0 μL of ddH2O. The amplification conditions were as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 0.5 min, 54°C for 0.5 min, and 72°C for 2.5 min, and a final extension for 5 min at 72°C. The amplicons of GtfAB were sequenced directly using Sanger’s DNA sequencing method.
3.4.6. Sequence Alignment and Function Prediction of GtfA and GtfB
Clustalw 2.0 software was used to perform multiple alignments of the sequences of GtfA and GtfB. Evolutionary divergence in these sequences was estimated using the maximum composite likelihood method in the MEGA4 software package. The neighbor-joining method was used to create a phylogenetic tree and infer evolutionary relationships. The Protein Homology/analogY Recognition Engine (PHYRE) program was used to search for structural homology. The GeneBank database was used to analyze conserved domains of GtfA and GtfB.
3.4.7. Construction of pACYCDuet-1, pETDuet-1, pGADT7, pGBKT7 and pGEX-5X-1 derivatives
Vector pACYCDuet-1(pAC) was used to express GtfA, GtfB, and GtfAB. First, GtfA, GtfB, and GtfAB were amplified from S. pneumoniae chromosomal DNA using PrimeSTAR Max DNA polymerase (TaKaRa Biotech Co., China) and corresponding primers (Appendix 1 in Supplementary File). The cleaved amplicons were ligated with vector pAC to construct pAC-A, pAC-B, and pAC-AB, respectively. Vector pETDuet-1(pET) was used to construct pET-P for the expression of PrsP1-374. The construction of pET-P was similar to that of pAC-A. All of the inserts included in pET and pAC were sequenced using Sanger’s DNA sequencing method.
The coding sequences for GtfA and GtfB were amplified from the extracted bacterial DNA of S. pneumoniae using PrimeSTAR Max DNA polymerase (TaKaRa Biotech Co., China) with primers GtfA EcoRI and BamHI, GtfB EcoRI and BamHI, respectively. PCR products were cleaved with EcoRI and BamHI, and cloned in frame into pGBKT7 (pBD) and pGADT7 (pAD) to construct pBD-A and -B and pAD-A and -B, respectively. Then, GtfA and GtfB were excised from pBD-A and -B by EcoRI and XhoI. The isolated fragments were subcloned in frame into vector pGEX-5X-1 (p5X) to construct p5X-A and p5X-B. All of the resulting constructs were confirmed using restriction enzyme digestion and DNA sequencing.
3.4.8. Co-Expression of Recombinant PsrP1-374 with GtfA or GtfB
Escherichia coli BL21 was co-transformed with the pET-P and pAC derivatives listed above. Bacteria were grown on LB agar plates supplemented with 100 μg/mL of ampicillin and 20 μg/mL of chloramphenicol. Bacteria harboring a different pAC derivative and a pET-P were cultured at 30°C to logarithmic growth phase at an OD600 of about 0.6 following added IPTG (final concentration 0.1 mM) to induce the expression of recombinant proteins (5). The bacterial lysates were analyzed by western blotting using monoclonal anti-His antibodies and polyclonal anti-S tag antibodies, respectively (5). Lectin blotting was used to investigate PsrP glycosylation in these bacterial lysates by using sWGA and four other agglutinins labeled with horseradish peroxidase-conjugates (5, 7).
3.4.9. Yeast two-Hybrid Analysis
Each pair of pAD and pBD derivatives was transformed into Saccharomyces cerevisiae AH109 using the LiAc Yeast Transformation Procedure (Clontech, USA). The transformed strain AH109 was grown on SD-LTHA agar plates for 48 hours to investigate the interaction between GtfA and GtfB.
3.4.10. Purification of GST-GtfA and -GtfB and In Vitro Translation of c-Myc- GtfA and -GtfB Proteins
Escherichia coli BL21 was cultured at 30°C to logarithmic growth phase at an OD600 of about 0.6, then IPTG (final concentration 0.1 mM) was added into the cultures to induce expression of recombinant proteins for 3 hours. Harvested bacterial pellets were used for the purification of GST-GtfA and -GtfB as previously described (6, 8). Supercoiled DNA with 1 μg of pBD-A and pBD-B was used to produce c-Myc-GtfA and -GtfB fusion proteins using the TnT® Quick Coupled Transcription/Translation System (Promega, USA) (6, 8).
3.4.11. Glutathione S-transferase Pull-Down Assays
The GST pull-down assays were carried out as previously described (6). The eluted proteins from GST beads were boiled for western blotting with a monoclonal anti-c-Myc antibody.
4. Results
4.1. Distribution of Pneumococcal Serotypes
A total of 141 pneumococcal isolates were collected for serotyping. Of these, 91 (64.54%) were successfully serotyped; 50 (35.46%) were non-typeable. Among the 91 clinical isolates, nine different S. pneumoniae serotypes were found (Appendix 3 in Supplementary File), and four pneumococcal isolates from sputum were identified as mixed serotypes; two contained 6A/B and 19A, one contained 19A and 19F, and one included 19A and 23F (Table 1). The most frequent serotypes were 19F (n = 42; 29.78%) and 19A (n = 12; 8.51%), followed by 6A/B (n = 10; 7.09%), 23F (n = 7; 4.96%), and 3 (n = 5; 3.55%; Table 1). These five serotypes together accounted for 53.89% of the isolates.
| Serotypes | No. (%) | PsrP (%) | GtfA (%) | GtfB (%) | PsrP-GtfA-GtfB (%) |
|---|---|---|---|---|---|
| 1 | 1 (0.07) | 0 | 0 | 0 | 0 |
| 3 | 5 (3.55) | 0 | 0 | 0 (0.00) | 0 |
| 6A/B | 10 (7.09) | 7 (70.00) | 7 (70.00) | 7 (70.00) | 7 (70.00) |
| 9V | 0 (0.00) | 0 | 0 | 0 | 0 |
| 10A | 4 (2.84) | 1 (25.00) | 1 (25.00) | 1 (25.00) | 1 (25.00) |
| 11A | 0 (0.00) | 0 | 0 | 0 | 0 |
| 14 | 4 (2.84) | 4 (100.00) | 4 (100.00) | 4 (100.00) | 4 (100.00) |
| 15A | 0 (0.00) | 0 | 0 | 0 | 0 |
| 15B/C | 2 (0.14) | 1 (50.00) | 1 (50.00) | 1 (50.00) | 1 (50.00) |
| 19A | 12 (8.51) | 6 (50.00) | 6 (50.00) | 6 (50.00) | 6 (50.00) |
| 19F | 42 (29.78) | 7 (16.67) | 7 (16.67) | 7 (16.67) | 7 (16.67) |
| 23F | 7 (4.96) | 5 (71.43) | 5 (71.43) | 5 (71.43) | 5 (71.43) |
| 6A/B+19A | 2 (0.14) | 2 (100.00) | 2 (100.00) | 2 (100.00) | 2 (100.00) |
| 19A+19F | 1 (0.07) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) |
| 19A+23F | 1 (0.07) | 1 (100.00) | 1 (100.00) | 1 (100.00) | 1 (100.00) |
| Non-typeable | 50 (35.46) | 16 (32.00) | 16 (32.00) | 16 (32.00) | 16 (32.00) |
| Total | 141 (100) | 51 (36.17) | 51 (36.17) | 51 (36.17) | 51 (36.17) |
The Prevalence of PsrP, GtfA and GtfB in Different Serotypes of Pneumococcal Isolates
4.2. Prevalence of GtfA, GtfB and PsrP
Of the 141 pneumococcal isolates in our collection, 51 (36.17%) were found to be GtfA-, GtfB-, and PsrP-positive (Appendix 4 in Supplementary File and Table 1). No isolate was found to be individually positive or negative for GtfA, GtfB, or PsrP. Among the nine serotypes we found, there were no GtfA-, GtfB- or PsrP-positive isolates of serotypes 1 and 3. The highest prevalence of GtfA, GtfB, and PsrP was found in isolates of serotype 14 (100.00%), followed by serotypes 23F (71.43%) and 6A/B (70.00%) (Table 1). Serotype 19F and the non-typeable isolates had 16.67% and 32.00% positive rates of GtfA, GtfB, and PsrP, respectively (Table 1).
4.3. Analysis of GtfA and GtfB Sequences
By searching the NCBI database, 34 complete genomic and chromosome sequences of the pneumococcal isolates were acquired, and the PsrP-secY2A2 island was only present in 16 of the 34 pneumococcal isolates. The GtfA and GtfB sequences of 23 pneumococcal isolates (16 from the NCBI database and 7 from clinical isolates presenting with different serotypes in this study) were used for multiple sequence assignment. The average homology of the GtfA sequences of TIGR4 and 22 other pneumococcal isolates was 99.59% with a maximum of 100.00% and a minimum of 98.41%, produced by S. pneumoniae UN030, a pneumococcal isolate identified to be serotype 19F. Thirteen kinds of variation were found in GtfA sequences between TIGR4 and 22 other pneumococcal isolates, and 8 kinds of variation (D270N, N283R, Q295P, D313E, H346R, L349I, D377N, and K454N) were found in GtfA sequences between TIGR4 and UN030. The phylogenetic tree showed that the sum of branch length was 0.044 (Appendix 5A in Supplementary File). These analyses suggest that the GtfA sequences of the pneumococcal isolates were conserved. We then inferred the function of the GtfA of the S. pneumoniae UN030 isolate. Conserved domain analysis revealed that GtfA was very closely related to the GT1 family of glycosyltransferase. The PHYRE program found significant structural similarity in the sequences of GtfA and Gtf1 of S. parasanguinis FW213 (Appendix 5B in Supplementary File).
The average homology of the GtfB sequences of TIGR4 and 22 other pneumococcal isolates was 98.43% with a maximum of 100.00% and a minimum of 95.07%, produced by S. pneumoniae 70585. Thirty-two kinds of variation were found in the GtfB sequences of TIGR4 and 22 other pneumococcal isolates, and 6 kinds of variation (G20V, A92V, T127K, P221R, C327R, and A332V) were found between TIGR4 and UN030. The sum of the branch length was inferred to be 0.09 using the neighbor-joining method, which was contributed mostly by SP70585 (Appendix 5C in Supplementary File). The GtfB sequences were also found to be conserved. Therefore, the GtfB of UN030 was used for function prediction. Conserved domain analysis showed that GtfB was most closely related to the GT1 family of glycosyltransferases. The PHYRE program found significant structural similarity in the sequences of GtfB and Gtf2 of S. parasanguinis FW213 (Appendix 5D in Supplementary File).
4.4. Involvement of GtfA and GtfB in the Glycosylation of Recombinant PsrP1-374
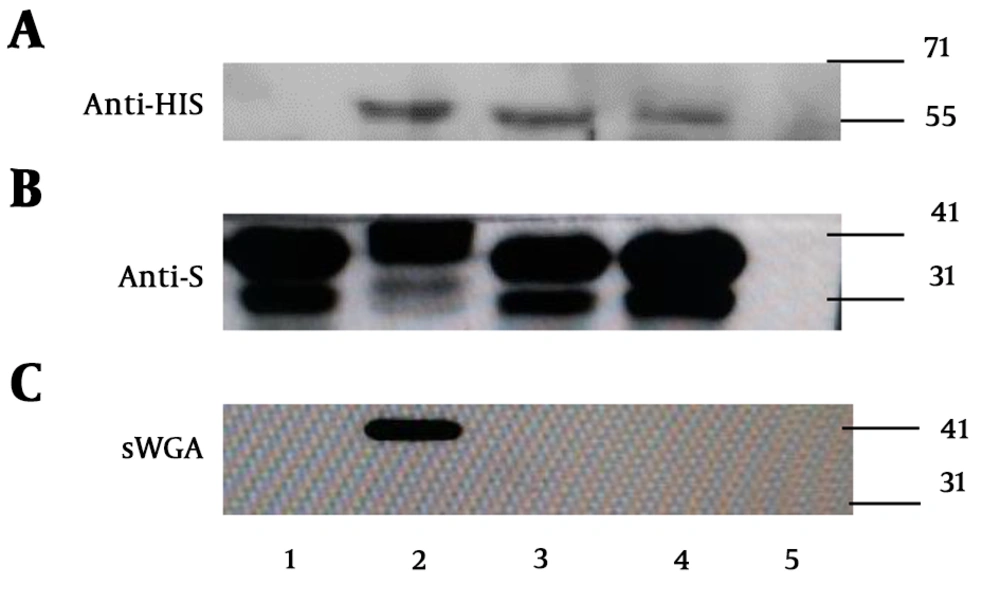
To investigate whether GtfA and GtfB of UN030 contribute to the glycosylation of PsrP, E. coli was used to construct an in vivo PsrP glycosylation system. Recombinant PsrP1-374 was used as the glycosyltransferase substrate, and GtfA and GtfB were used as the glycosyltransferases. In this system, the individual expression of PsrP1-374 or co-expression of PsrP1-374 and GtfA or GtfB led to the production of a PsrP1-374 doublet when probed with anti-S tag antibody (Figure 1B, lanes 1, 3, and 4). The co-expression of GtfA, GtfB, and PsrP1-374 produced a PsrP1-374 doublet of higher molecular weight and slower migration rate (Figure 1B, lane 2). The PsrP1-374 band with the slower migration rate bound to N-acetylglucosamine (GlcNAc)-specific lectin sWGA (Figure 1C, lane 2) rather than any of the four other lectins, including Ulex europaeus agglutinin I, peanut agglutinin, Dolichos biflorus agglutinin, and concanavalin A, which bind specifically to fucose, galactose, N-acetylgalactosamine, and glucose (or mannose), respectively. It was concluded that PsrP1-374 was only glycosylated by the GlcNAc moieties in E. coli. This finding verifies that both GtfA and GtfB of UN030 are glycosyltransferases and required for the glycosylation of recombinant PsrP1-374 in the E. coli glycosylation system.
Analyses of PsrP1-374 expression in an E. coli glycosylation system. E. coli strains were transformed with plasmids pET-P and pAC (lane 1), pET-P and pAC-AB(lane 2), pET-P and pAC-B (lane 3) and pET-P and pAC-A (lane 4), pET and pAC (lane 5) and subjected to immunoblot analysis with A, Anti-HIS tag monoclonal antibody; B, Anti-S tag polyclonal antibody; C, GlcNAc-reactive lectin sWGA.
4.5. GtfA Interaction with GtfB
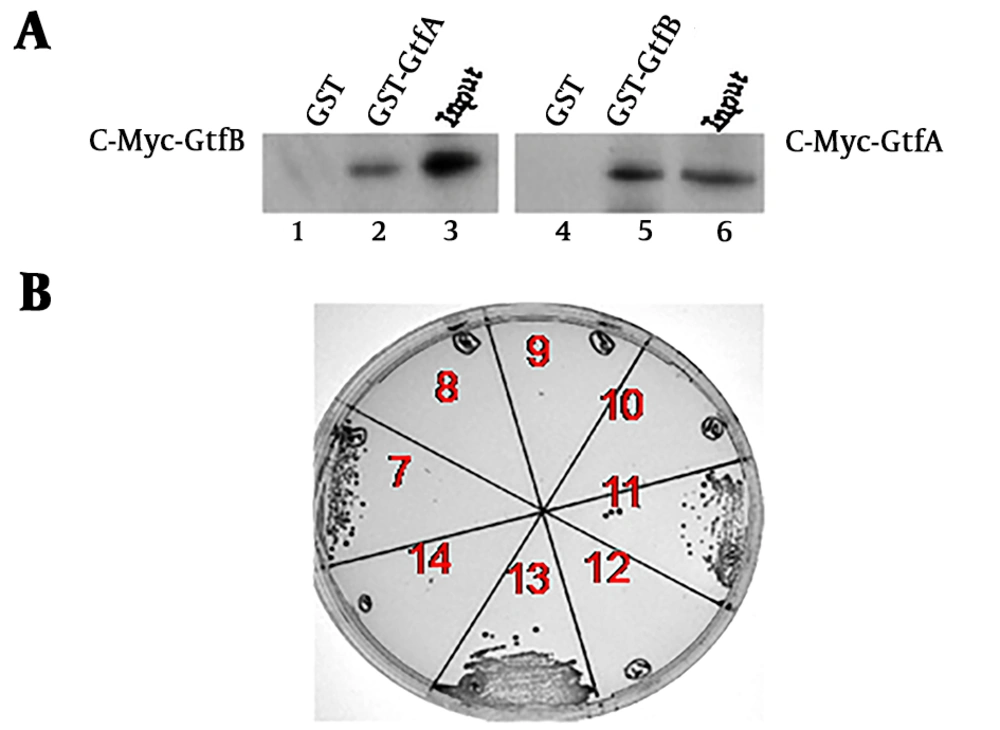
As only co-expression of GtfA and GtfB led to the glycosylation of PsrP1-374, it was reasonable to speculate that GtfA might form an enzyme complex with GtfB to regulate PsrP glycosylation. Matchmaker Yeast Two-Hybrid System 3 (Clontech, USA) was used to determine the interaction between GtfA and GtfB. Like the positive control (Figure 2A, section 13), numerous colonies were grown on an SD-LTHA agar plate when AH109 was co-transformed with pAD-GtfA and pBD-GtfB or pAD-GtfB and pBD-GtfA (Figure 2A, sections 7 and 11). The other co-transformants created few colonies or no colony on the same agar plate (Figure 2A, sections 8, 9, 10, and 12), similar to the negative control (Figure 2A, section 14). GST pull-down assays were then used to determine the interaction between GtfA and GtfB. GST-GtfA and -GtfB bound to c-Myc-GtfB (Figure 2B, lane 2) and -GtfA (Figure 2B, lane 5), respectively, whereas GST had no ability to bind to either c-Myc-GtfA (Figure 2B, lane 4) or c-Myc-GtfB (Figure 2B, lane 1). These findings indicate that GtfA interacted directly with GtfB, resulting in an enzyme complex.
5. Discussion
In this study, GtfA, GtfB, and PsrP were found in 36.17% of the collected S. pneumoniae isolates, which is different from a recent finding that PsrP was present in 51.7% of invasive S. pneumoniae isolates and in 48.3% of carrier S. pneumoniae isolates (15), indicating there are regional differences in the prevalence of GtfA, GtfB, and PsrP. Because GtfA, GtfB, and PsrP are not present in about 50% of clinical isolates of S. pneumoniae, a PsrP-SecY2A2 island is not essential for S. pneumoniae growth. Nine serotypes were identified in 91 clinical isolates and the most frequent serotypes were 19F, 19A, 6A/B, and 23F, similar to the recently reported findings (12). The highest prevalence of PsrP was found in serotype 14, whereas no PsrP-positive isolate was found in serotypes 1 and 3 in the present study. Selva et al. reported that PsrP was present in 80.15% and 9.52% of isolates of serotype 1 and serotype 3, respectively (15). In this study, only 6 isolates were identified as serotype 1 or 3, indicating that an insufficient number of isolates caused PsrP-negative findings in serotypes 1 and 3. Therefore it is concluded that the presence of a PsrP-SecY2A2 island is not associated with a certain serotype.
Mature PsrP was demonstrated to bind to sWGA, suggesting it was glycosylated by N-acetylglucosamine (GlcNAc) moieties (2, 3). Glycosyltransferase A and GtfB have been shown to be required for the glycosylation of PsrP in S. pneumoniae serotype 4, strain TIGR4 (2). In the present study, GtfA and GtfB were found to be present in 7 of 9 serotypes of S. pneumoniae, and the average homology of the sequences of GtfA and GtfB was 99.59% and 98.53%, respectively, suggesting that GtfA and GtfB are conserved. Phylogenetic tree analysis of GtfA sequences showed that pneumococcal strain TIGR4 was most distantly related to serotype 19F, isolate UN030. Moreover, serotype 19F was the most frequent in the present study, thus, GtfA and GtfB of UN030 were used for further experiments. A significant structural similarity between GtfA and Gtf1, as well as GtfB and glycosyltransferase-stabilizing protein Gtf2 was revealed. Conserved domains identification showed that both GtfA and GtfB were most closely related to the GT1 family of glycosyltransferases. Therefore, it is reasonable to conclude that GtfA and GtfB of UN030 are glycosyltransferases.
To verify the deduction that GtfA and GtfB of UN030 are glycosyltransferases, E. coli served as a scaffold for constructing an in vivo PsrP glycosylation model. The expression of S-tag-fused PsrP1-374 in E. coli was probed with anti-S-tag polyclonal antibodies. Co-expression of PsrP1-374 with GtfA and GtfB in E. coli resulted in two bands with higher molecular weight and slower migration rate. Only the PsrP1-374 band at the ~50-kD position with a slower migration rate was capable of binding to sWGA. These results demonstrate that GtfA and GtfB are glycosyltransferases that transfer GlcNAc moieties to the peptide backbone of PsrP in S. pneumoniae strain UN030, which strongly supports the previous findings that mature PsrP is glycosylated by GlcNAc, and GtfA and GtfB are required for transferring GlcNAc to the peptide backbone of PsrP in S. pneumoniae strain TIGR4 (2, 3).
In the present study, neither GtfA nor GtfB had the ability to transfer GlcNAc moieties to the peptide backbone of PsrP, as probed with lectin sWGA. Therefore, it is reasonable to deduce that GtfA interacts with GtfB and forms the glycosyltransferase complex GtfA-GtfB, which is required for the glycosylation of PsrP. GtfA interaction with GtfB was demonstrated using the in vitro GST pull-down assays and yeast two-hybrid analyses in the present study. Supporting these results, yeast two hybridization and GST pull-down also revealed that Gtf1 interacted with Gtf2 in S. parasanguinis FW213. Moreover, a significant structural similarity was found between GtfA and Gtf1, as well as GtfB and Gtf2 (7, 9). Therefore, it is concluded that GtfA interacts with GtfB and forms the enzyme complex GtfA-GtfB with GtfB, which is required for PsrP glycosylation in S. pneumoniae. In addition, these findings also explain why the activity of the mixture of GtfA and GtfB was 100-fold higher than that of the individual GtfA (10).
5.1. Conclusions
In sum, nine different S. pneumoniae serotypes were found in the present study, and the most frequent serotype was 19F. The prevalence of GtfA, GtfB, and PsrP was 36.17% in the collected S. pneumoniae isolates, and it is not associated with a particular serotype. Glycosyltransferase A and GtfB are conserved in the collected isolates of S. pneumoniae. Glycosyltransferase A interacts with GtfB, forming the glycosyltransferase complex GtfA-GtfB, and it is required for PsrP glycosylation.