1. Background
The ability of Acinetobacter to survive in moist and dry environments allows frequent contamination of the hospital environment with Acinetobacter species (1) because the organism can survive for long times in dry conditions (2) so that dry wards of the hospital settings are potential reservoirs of the infection (3). Lately, the bacteria of the genus Acinetobacter emerged as a leading cause of nosocomial infections and Acinetobacter baumannii accounted for nearly 80% of the all reported Acinetobacter infections (4), affecting mainly immunocompromised patients, especially those who experienced long periods of hospital stay (5). Nowadays, an arising problem with A. baumannii is acquiring resistance to almost all commonly used antibiotics including aminoglycosides fluoroquinolones and ß-lactam agents (4, 6) and remaining susceptible only to older, more toxic agents, leaving limited and suboptimal choices for treatment (7). The worldwide emergence of multidrug-resistant bacteria (MDR) is endangering the therapeutic efficacy of antibiotics (3, 4). A major concern while treating these infections is the proper choice of antibiotics as improper therapy will affect the patient health (8).
Aminoglycosides are a group of antibiotics that are characterized by the presence of an aminocyclitol ring linked to amino-sugars in their structure. They are most frequently used in the treatment of invasive infections produced by non-lactose fermenters such as A. baumannii and Pseudomonas aeruginosa (9). Aminoglycoside resistance is mainly due to the presence of enzymes including Aminoglycoside Acetyltransferases (AACs), Aminoglycoside Phosphotransferases (APTs), and Aminoglycoside Nucleotidyltransferases (ANTs) that catalyze the chemical modification of the antibiotic by acetylation, phosphorylation, and adenylation or participate in 16S rRNA methylation (10, 11). Another mechanism responsible for aminoglycosides resistance is a decrease in the drug uptake due to either outer membrane impermeability or overexpression of active efflux pumps (11). Recently, aminoglycosides have served as substrates for several multidrug efflux pumps including five superfamilies of bacterial transporters (12).
The resistance nodulation cell division (RND) superfamily plays an important role in the resistance of Gram-negative bacteria (13, 14). Experimental studies indicate that the AdeABC efflux pump in A. baumannii is responsible for developing resistance to aminoglycosides (15, 16). AdeABC efflux pumps are the members of the RND superfamily localized in the cytoplasmic membrane incorporated with the membrane fusion protein (MFP) and the outer membrane protein (OMP) (16, 17), which helps the bacteria to export substrates directly out of the cell, using proton-motive force as an energy source (18). This decreases the intracellular concentration of the drug and makes the bacteria more resistant (16). Overexpression of the AdeABC has been correlated with decreased susceptibility to aminoglycosides (15, 16). The minimum inhibitory concentrations (MICs) are usually 2 to 8 folds higher in strains overexpressing an efflux pump than in other susceptible strains of the same species (19, 20). Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) is a mitochondrial proton gradient uncoupler that is used for the detection of different types of efflux pumps and it decreases the resistance of multi-drug resistant (MDR) bacteria (21). Pandrug resistance has been variously defined as a non-susceptibility to all commercial antimicrobial agents in the same class (22). The emergence of pan-aminoglycoside-resistant Gram-negative bacteria has been increasingly reported in recent years (23).
2. Objectives
In our study, we aimed to phenotypically detect the presence of efflux mechanism using CCCP in aminoglycoside resistant clinical A. baumannii strains isolated from different hospital wards.
3. Methods
3.1. Ethics Statement
All experiments in this study were conducted with the approval of the ethics committee of Beni-Suef University, Egypt MI (1046).
3.2. Samples and Bacterial Strains
A total of 57 A. baumannii non-duplicated samples were collected in a period between April 2014 and January 2015 from patients with clinically diagnosed respiratory tract infections, urinary tract infections, skin and soft tissue infections, wound infections, bacteremia, bacterial peritonitis and meningitis at two Egyptian hospitals: Hospital A, El Demerdash Hospital (Cairo, Egypt) and hospital B, El Mansoura Teaching Hospital (Dakahlia, Egypt). The average age of the patients was 48. This study was conducted using clinical specimens from those cases that developed their infections after hospital admission by 48 hours. Samples were promptly transferred to the laboratory and each sample was inoculated on nutrient agar (Oxoid, UK), McConkey agar (Oxoid, UK) and blood agar (Oxoid, UK) and incubated according to the approved microbiological procedures. Bacterial isolates were identified as A. baumannii based on the standard biochemical tests including citrate test, Catalase test, oxidase, Indole, oxidative fermentation test, fermentation of galactose, glucose, lactate, malonate, maltose, mannose, rhamnose, and xylose. arginine dihydrolase, histidine, leucine, malate, phenylalanine deaminase, and tyrosine hydrolysis tests also were carried out (24), followed by PCR amplification of blaOXA-51-like gene.
3.3. Molecular Identification of A. baumannii Isolates Using blaOXA-51-like Gene
DNA extraction for PCR reactions was performed according to the method described in (25) and molecular identification of the blaOXA-51-like gene, which is an intrinsic carbapenemase gene found only in A. baumannii, was performed using PCR according to the method described in (26).
3.4. Bacterial Susceptibility Testing
Antibiotic susceptibility testing against A. baumannii isolates was carried out according to the Kirby-Bauer disk diffusion method using the following antibiotics; amikacin (30 µg), gentamicin (30 µg), kanamycin (30 µg), neomycin (30 µg), netilmicin (10 µg), streptomycin (300 µg), and tobramycin (30 µg) all supplied from (Oxoid, UK). A. baumannii ATCC 17978 and P. aeruginosa ATCC 27853 were used as the reference strains. The results were interpreted according to CLSI (27, 28). The isolates were classified as pan-aminoglycoside resistant according to Magiorakos et al. 2012 (22). The minimum inhibitory concentration (MIC) was determined using the microdilution broth method (29, 30). Each stock solution of the antimicrobial chemotherapeutic agent was prepared from the authentic powder supplied by respective manufacturers. Gentamicin and neomycin were gifted from Memphis pharmaceutical Co. (Cairo-Egypt). Tobramycin was gifted from El-Kahera pharmaceutical Co. (Cairo-Egypt). Amikacin was purchased as Amikin Vial from Bristol Myers Squibb (Princeton, New Jersey) and kanamycin was supplied from Schering Plough, Kenilworth (New Jersey, USA).
3.5. Studying the Effect of Efflux Pump Inhibitor on Antibiotic Susceptibility
In each Muller Hinton agar plate, containing 2-1024 µg/mL of each tested antibiotic (amikacin, gentamicin, kanamycin, neomycin, and tobramycin), the efflux pump inhibitor CCCP supplied from Sigma (St. Louis, MO) was added at a final concentration of 12.5 µM. Then, the MIC was determined again. A plate containing only CCCP was used as the control. A reduction of at least two folds in the MIC after addition of the CCCP was the positive indication for the existence of efflux pump in the isolates (31).
4. Results
4.1. Bacterial Isolates and Aminoglycoside Resistance
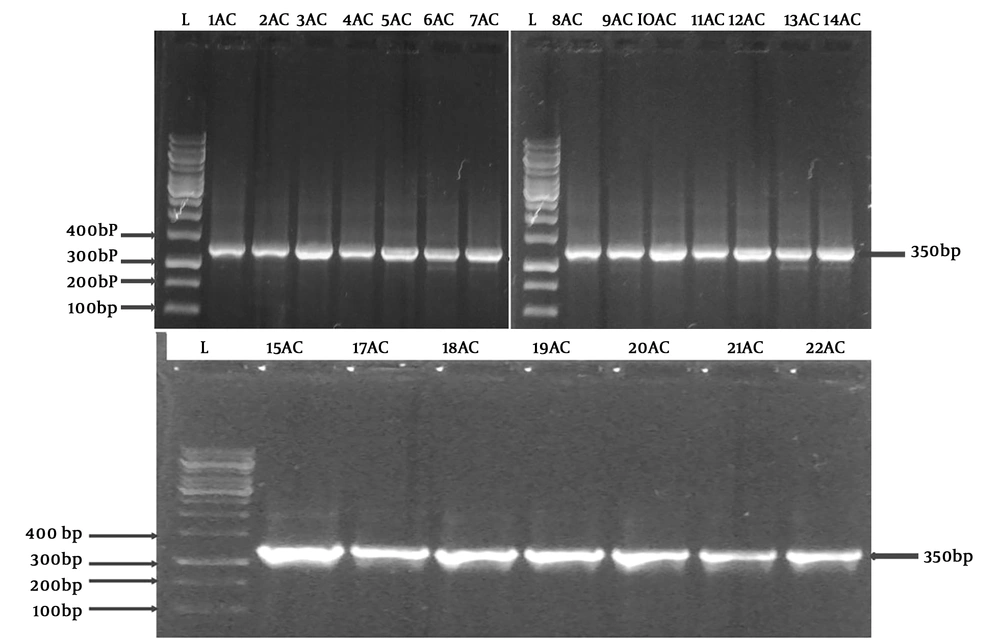
In a nine-month period, 57 isolates of A. baumannii were isolated. Thirty nine strains from hospital A and 18 from hospital B were collected from hospitalized patients and their infections were classified as follows: Skin and soft tissue infections (28%), respiratory tract infections (24.6%), bacteremia (24.6%), urinary tract infections (10.5%), meningitis (7.1%), wound infections (3.5%), and bacterial peritonitis (1.7%). All Acinetobacter isolates were positive for catalase, citrate, and oxidative fermentation. All tubes gave a yellow color in the aerobic tube and a green color in the anaerobic tube; all isolates were able to ferment galactose, glucose, lactate, malonate, maltose, mannose, rhamnose, and xylose; in addition, arginine dihydrolase, histidine, leucine, malate, phenylalanine deaminase, and tyrosine hydrolysis were positive. The blaOXA-51 gene was present in all the isolates. A representative agarose showing the PCR products is represented in Figure 1. High resistance levels were reported to tobramycin, amikacin, kanamycin, neomycin, streptomycin, and gentamicin. The MIC breakpoints recommended by CLSI (28, 30) against A. baumannii isolates were ≥ 16 µg/mL for gentamicin and tobramycin while for amikacin, kanamycin, and neomycin, they were ≥ 64 µg/mL for the isolates to be considered as resistant. The MICs range of A. baumannii was from 2 to 1024 µg/mL as summarized in Table 1 based on the tested aminoglycosides. 89.4%, 84.3%, 68.5%, 57.8%, and 49.1% of the isolates represented the resistance range of the MICs for kanamycin, tobramycin, amikacin, gentamicin, and neomycin, respectively.
Detection of blaOXA-51 gene using PCR. A electrograph of a representative agarose gel (1.2% wt/vol %) showing the PCR products. L refers to a100 bp DNA size marker ladder while the amplified fragments of A. baumannii isolates appeared in other lanes. The amplicons showed up on the predicted size of the blaOXA-51 gene (350 p).
| Antibiotic MIC | Value, Percentage |
|---|---|
| Gentamicin | |
| S | 42.2 |
| I | 0 |
| R | 57.8 |
| Kanamycin | |
| S | 10.6 |
| I | 0 |
| R | 89.4 |
| Tobramycin | |
| S | 7 |
| I | 8.7 |
| R | 84.3 |
| Neomycin | |
| S | 36.7 |
| I | 14.2 |
| R | 49.1 |
| Amikacin | |
| S | 26.2 |
| I | 5.3 |
| R | 68.5 |
Abbreviations: I, intermediately resistant; R, resistant; S, sensitive.
4.2. Effects of CCCP on Aminoglycoside Resistance
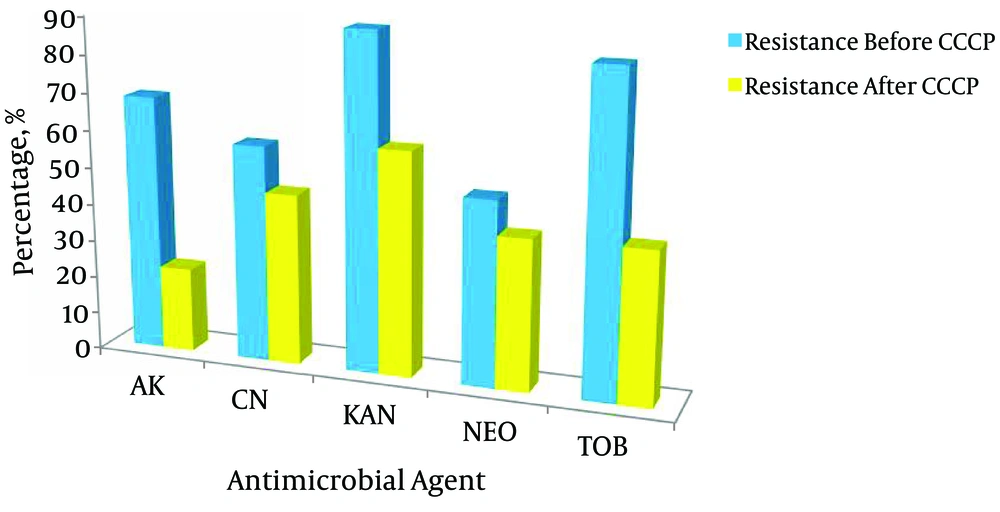
In order to determine the role of the efflux pump in aminoglycoside resistant isolates, we evaluated the MIC of the tested antibiotics in the presence of 12.5 µM CCCP and then compared them with the MIC without the efflux pump inhibitor (EPI). The results indicated that 19.4% of the isolates became less resistant to kanamycin, 44% to tobramycin, and 46% to amikacin but lower rates were recorded against gentamicin and neomycin as 12.2% and 9.4%, respectively. All the results are summarized in Table 2 and Figure 2. A two-fold or greater reduction in the MIC values of the antimicrobial agent with CCCP versus the antibiotic alone was the criterion for significance (21, 28, 30). All isolates grew in the presence of CCCP alone, indicating that the concentration of 12.5 µM did not have any antibacterial effect on the tested isolates.
| Fold Reduction | Gentamicin | Kanamycin | Tobramycin | Neomycin | Amikacin |
|---|---|---|---|---|---|
| 512 folds | - | - | - | - | - |
| 256 folds | - | - | - | - | - |
| 128 folds | 9 | 10 | 8 | 25 | 8 |
| 64 folds | 6 | 8 | - | 21 | 5 |
| 32 folds | - | - | 40 | 14 | 36 |
| 16 folds | 3 | 2 | 4 | - | 26 |
| 8 folds | 9 | 2 | 10 | 11 | - |
| 4 folds | - | 2 | 32 | 21 | 10 |
| 2 folds | 3 | - | 6 | 8 | 15 |
5. Discussion
Acinetobacter baumannii is a Gram-negative non-lactose fermenting coccobacilli causing nosocomial infections such as skin and soft tissue infections, urinary tract infections, bloodstream infections, and pneumonia. The tendency of A. baumannii to acquire resistance to most commonly used antibiotics contributes to a major public health problem (21, 32, 33). Multidrug and pan-drug-resistant isolates are now arising in hospital settings. Both antibiotic resistance and the ability to persist for long periods in hospital environments are responsible for epidemics caused by this organism (16). Many reports confirmed A. baumannii is difficult to treat due to several virulence factors and resistance genes conferring to high levels of resistance (34).
Evaluation of the sensitivity patterns of different isolated A. baumannii strains against most frequently used aminoglycosides in our hospitals was necessary to get a better understanding of their behavior. The highest resistance levels in A. baumannii were recorded against streptomycin (95%), followed by kanamycin (87%), amikacin (86%), tobramycin (81%), neomycin (70%), netilmicin (63%), and gentamicin (51%). In Korean hospitals (35), it was reported that A. baumannii isolates were highly resistant to amikacin, gentamicin, streptomycin, and tobramycin with resistance rates of 85%, 75%, 92%, and 79%, respectively. Our results were nearly consistent with the results of another Egyptian study performed in 2012 in which all Acinetobacter isolates were resistant to kanamycin, amikacin, and tobramycin and 50% were resistant to gentamicin, neomycin, and netilmicin (36).
A study in Kenya tested antibiotic susceptibility to seven aminoglycosides and reported the resistance rates of the isolates to streptomycin as 87%, kanamycin as 81%, amikacin as 46%, tobramycin as 65%, neomycin as 62%, and gentamicin as 69%. This study corroborated well with our results with the exception of amikacin (37). Different antimicrobial susceptibility studies conducted in Iran on A. baumannii isolates showed among all aminoglycosides tested, netilmicin had the highest susceptibility rate (38.7%), followed by tobramycin (30.7%), amikacin (18.6%), gentamicin (8.0%), and kanamycin (6.7%) (38). Another study in 2014 stated that the maximum resistance was against streptomycin (90%), gentamicin (83%), tobramycin (83%), and kanamycin (80%) (39). Aminoglycoside resistance in 87 A. baumannii strains was evaluated, indicating that netilmicin had the highest activity (67.8%) against the isolates. The resistance rates for kanamycin, gentamicin, amikacin, and tobramycin were 87.3%, 57.4%, 54%, and 47.1%, respectively (40). The discrepancy in these results could be related to differences in antimicrobial therapies, patterns of antibiotic resistance, quality of surveillance programs, geographic conditions, and environmental factors of different countries.
All living cells express different efflux transporters, which protect them from the toxic effects of organic chemicals (41). Increased efflux of antibiotics from the bacterium lowers the drug accumulation rate, which augments the MIC values (41). It has been found that inhibitors of the efflux pumps reverse the multidrug resistance in A. baumannii and other Gram-negative bacteria (19). The antimicrobial susceptibility with and without carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was studied and the results indicated that 19.4% of the isolates became less resistant to kanamycin, 44% to tobramycin, and 46% amikacin while lower rates were recorded against gentamicin and neomycin (12.2% and 9.4%, respectively). A significant reduction in their antimicrobial activity was observed from two to eight folds or more, which indicates that the efflux mechanism plays an important role in aminoglycoside resistance.
A study in Iran reported that among 80 A. baumannii strains, 90% (72/80) were resistant to amikacin and gentamicin; upon addition of 25 µg/mL CCCP, a reduction in the minimum inhibitory concentrations from 4 to 64 folds occurred in 25% of the isolates (42). A study in Thailand examined amikacin, gentamicin, kanamycin, and neomycin against 100 A. baumannii isolates and reported resistance rates of 80%, 90%, 90%, and 86%, respectively. The MIC values were observed as 64 µg/ml for amikacin and kanamycin and 16 µg/ml for gentamicin and neomycin. After addition of 25 µg/ml carbonyl cyanide m-chlorophenylhydrazone, a reduction in the susceptibility levels of the isolates was observed as 47% to amikacin, 69% to gentamicin, 75% to kanamycin, and 66% to neomycin. In addition, the results revealed that at least a 4-fold reduction was observed in 40% of the amikacin, 62% of the gentamicin, 32% of the kanamycin, and 47% of the neomycin resistant isolates. This suggested that the high resistance levels were probably contributed to the active efflux pumps (43).
Rajamohan and coworkers tested the susceptibility levels of 83 resistant A. baumannii isolates at two different hospitals in Ohio against amikacin, streptomycin, gentamicin, and kanamycin. They also studied the effect of the efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) on the MIC values of the isolates and found a drastic reduction in the MIC values of 66/83 isolates, which indicated efflux mechanism plays an important role in aminoglycoside resistance (44). In concordance with our results, a study conducted at five hospitals in Taiwan to investigate efflux pumps in 21 multidrug resistance A. baumannii clinical isolates and their relationship with the presence of efflux pump inhibitors such as carbonyl cyanide 3-chlorophenylhydrazone found a 4-fold reduction in the MIC values of amikacin and gentamicin (45). The First Hospital of China Medical University in a period between January 2009 and December 2010 determined the antimicrobial susceptibility of amikacin and netilmicin against A. baumannii strains and revealed 11 were sensitive and 10 were resistant to amikacin while 15 were sensitive and 14 were resistant to netilmicin; the MIC values of amikacin and netilmicin were 512 µg/mL and 256 µg/mL, respectively. The detection of the efflux phenotype using CCCP at a final concentration 10 µg/mL decreased the values of the MIC by two folds for the sensitive stains and by four folds or more for the resistant strains (46).
5.1. Conclusions
In conclusion, it is obvious from our study and other international studies that A. baumannii isolates in hospital infections have been increasing in recent years. Furthermore, the results suggest the role of efflux pumps in conferring resistance to aminoglycosides in Acinetobacter isolates. Efflux pump inhibitors provide a new horizon to combat bacterial resistance together with antibiotics, and this idea is finding more and more supporters lately. Researchers are advised to produce new safe and effective efflux pump inhibitors to be used clinically.