1. Background
Candida parapsilosis is one of the most common yeasts, which is responsible for invasive candidiasis (1). It is considered as a nosocomial yeast pathogen that causes a high variety of clinical manifestations such as endocarditis, fungemia, septic arthritis, pancreatitis, endophthalmitis, and meningitis (2). Since the early 1990s, based on a variety of molecular studies such as randomly amplified polymorphic DNA analysis (RAPD) (3), mitochondrial DNA sequences analysis (4), and internal transcribed spacer (ITS) sequencing (5), C. parapsilosis has been categorized as a complex consisting of three genetically distinct groups (named I, II and III). Tavanti et al. replaced the groups II and III with C. orthopsilosis and C.metapsilosis based on multilocus sequence typing (MLST) data and identified three separate species as the C. parapsilosis complex (6). Restriction enzyme BanI was used for the differentiation of these three new species by digestion of the polymerase chain reaction (PCR)-amplified product of the secondary alcohol dehydrogenase-encoding (SADH) gene (1). The restriction enzyme BanI can digest amplicons to generate different fragments as two fragments for C. parapsilosis sensu stricto, four fragments for C. metapsilosis, and no restriction site for C. orthopsilosis (6). In Iran, Mirhendi et al. used the SADH gene and NlaIII enzyme for identification of the C. parapsilosis species complex (1).
Many pathogenic microbes possess constitutive and inducible hydrolytic enzymes that assist the invasion of host tissues and destroy membrane integrity, leading to disruption of host structures. An important virulence factor in Candida species is the secretion of extracellular hydrolytic enzymes, which contribute to host-tissue invasion by digesting proteins (7). Candida spp. produce a large variety of secreted hydrolases. In this regard, phospholipase and proteinase are the two most important hydrolytic enzymes. Extracellular aspartyl proteinase (Sap) is a putative virulence factor in Candida species (8). In our previous study, Candida species were isolated from onychomycosis and oral lesions of diabetic and denture stomatitis patients. These species were identified as C. parapsilosis by a conventional and PCR-restriction fragment length polymorphism (RFLP) method using MspI restriction enzyme for the digestion of ITS amplified fragments (9-11).
2. Objectives
This study was conducted to discriminate the C. parapsilosis species complex among clinical C. parapsilosis isolates using the digestion of SADH gene fragment by BanI restriction enzyme and evaluation of aspartyl proteinase activity as one of the potential virulence factors in the pathogenesis of the isolates.
3. Methods
3.1. Ethics Statement
The Ethics Committee of Departmental Review Board of Shiraz University of Medical Sciences approved this study (ethical code: IR.SUMS.REC.1393.S7000).
3.2. Isolates
In this study, 71 species of C. parapsilosis were collected from clinical samples (i.e., nail and oral lesions of diabetic and denture stomatitis patients) of Department of Parasitology and Mycology of Shiraz School of Medicine, Iran. Previously, these isolates were identified to the species level as Candida parapsilosis sensu lato by the amplification of ITS region and using the restriction fragment length polymorphism (RFLP)-MspI method (9-11). The samples were subcultured on Sabouraud dextrose agar (Merck, Germany) and incubated at 30°C for 24 hours before use.
3.3. DNA Extraction
According to the method proposed by Makimura et al. (12), genomic DNA of C. parapsilosis isolates was extracted with some modifications. Briefly, a small amount of the colonies was suspended in 100 µL of lysis buffer (i.e., 100 mM Tris-HCl, 0.5% SDS, and 30 mM EDTA) and boiled for 15 minutes at 100°C. Then, 2.5 M potassium acetate solution was added and kept on ice for 1 hour. The tubes were then centrifuged at 12,000 rpm for 5 minutes, and the supernatant was separated and precipitated with isopropanol. The yeast DNA in the supernatant was washed with ethanol twice and air dried. The remaining solution was re-suspended in 50 µL of distilled water. The purity and quantity of DNA were evaluated by electrophoresis on 1.2% agarose gel with an absorbance ratio of A260/A280 and nano-drop reading, respectively.
3.4. PCR-RFLP
The SADH gene was amplified by PCR using S1F as forward (5’-GTTGAT GCTGTTGGATTGT-3’) and S1R as reverse (5’-CAA-TGC-CAA-ATC-TCC-TAT-GC-3’) primers (Fermentas, Lithuania) to produce approximately 720-bp fragments (6). Amplification was performed in final 25 µL volumes, containing 12.5 µL of Master Mix (Amplicon, UK), 0.5 µL of S1F and S1R primers, 2 µL of genomic DNA, and 9.5 µL of dH2O. The PCR condition was as follows: the first step at 94°C for 3 minutes, the second step (35 cycles) at 94°C for 30 seconds, 51°C for 30 seconds, and 72°C for 30 seconds, and a final step at 72°C for 5 minutes. Restriction enzyme BanI (New England Biolabs, UK) digested the PCR product at 37°C for 120 minutes (Thermo, Lithuania) using 0.5 µL (5 U) of enzyme, 1.5 µL of buffers, 5 µL of PCR product, and 8 µL of molecular grade water. After digestion, the RFLP patterns for the discrimination of three species of the C. parapsilosis complex were obtained: two fragments of 521 and 196 bp for C. parapsilosis sensu stricto, four fragments of 370, 188, 93, and 60 bp for C. metapsilosis, and no restriction site (721 bp) for C. orthopsilosis. Strains C. parapsilosis sensu stricto ATCC 4344 and C. metapsilosis (professor Mirhendi, University of Isfahan) were used as positive controls and C. albicans ATCC10261 was used as the negative control.
3.5. Determination of Aspartyl Proteinase Activity
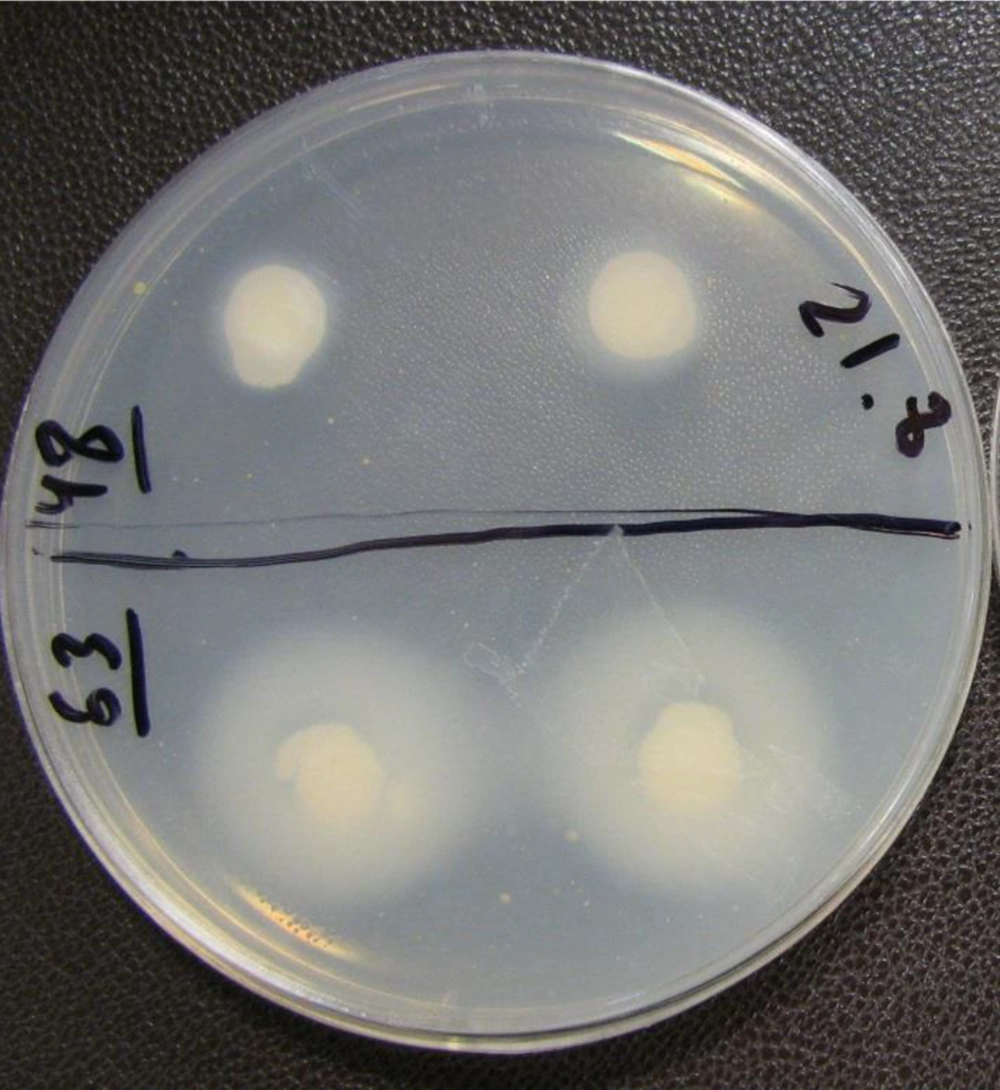
All the isolates were evaluated for determining their ability to secrete aspartyl proteinase on media containing bovine serum albumin (BSA) (13). A suspension with a density of 107 yeasts/ mL in yeast extract peptone dextrose (YEPD) medium (Difco, USA) was prepared and 6 mm-filter paper disks were dipped into the suspension and applied to the plates. The plates were incubated at 30°C for six days. The opaqueness of the agar around the disks (zone of proteolysis) presented degradation of the protein. The millimetric precipitation zone (PZ) of the opacity around the disks was measured and expressed as negative (-) for no clearance, positive (+) for mild, double positive (++) for moderate, triple positive (+++) for strong, and four positives (++++) for very strong enzyme activity. The standard strain was used as positive control, and each strain was analyzed in duplicate.
3.6. Statistical Analysis
Fisher’s exact test was run to analyze the data, and P value less than 0.05 was considered statistically significant.
4. Results
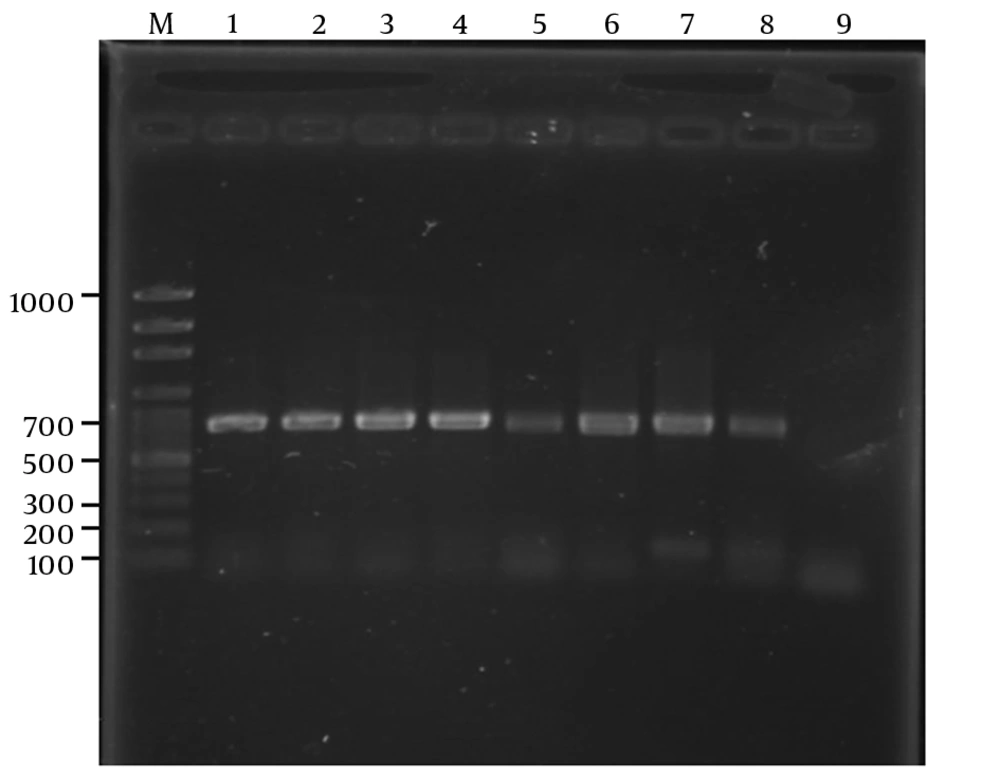
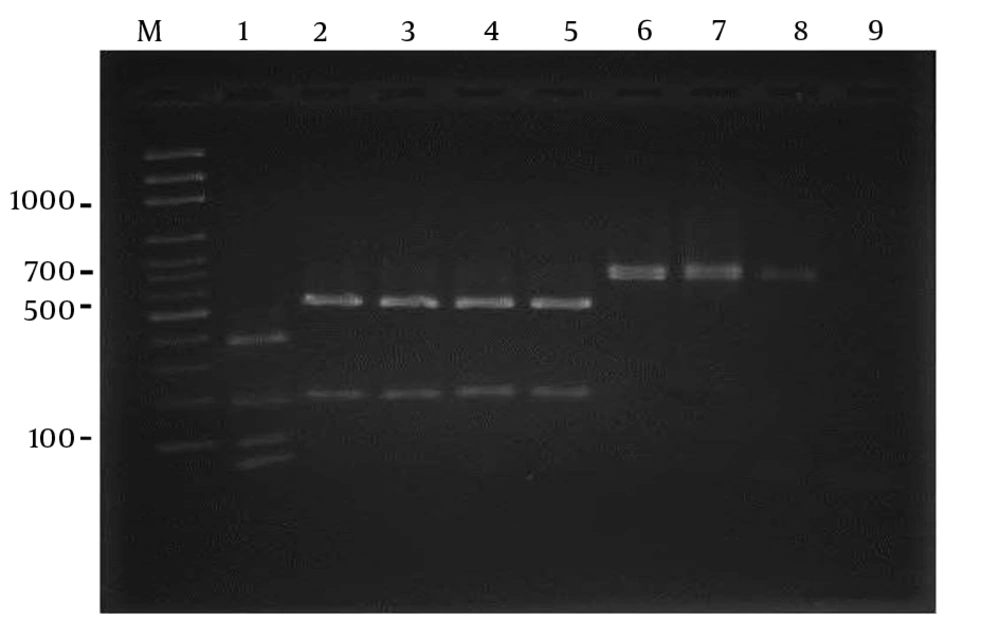
Using S1F and S1R primers, the SADH gene was successfully amplified and all the 71 isolates showed a single band of approximately 720 bp (Figure 1). In this study, using the restriction enzyme Ban1, 65 (91.5%) isolates were identified as C. parapsilosis sensu stricto, while 6 (8.5%) were proved to be C. orthopsilosis (Figure 2). Candida metapsilosis was not identified in this study.
RFLP pattern of PCR products obtained from Candida parapsilosis sensu lato strains after digestion with BanI restriction enzyme. Lane M: molecular size marker 100-bp. lane1: Candida metapsilosis (standard), lane 2: Candida parapsilosissensu stricto (standard): lanes 3 - 5 clinical samples; lanes 6–8: Candida orthopsilosis (clinical samples); lane 9: negative contro.
Of the 65 isolates of C. parapsilosis sensu stricto, 46 (70.76%) were proteinase producers (Figure 3), most of which (21/65) presented a very strong enzymatic activity. The rest of the isolates showed proteinase activity as follows: 19 (29.2%) strong, 3 (4.6%) mild, 3 (4.6%) weak positive, and 19 (29.23%) negative.
For C. orthopsilosis, 5 out of 6 (83.2%) isolates were able to produce proteinase, and 4 (66.6%) isolates showed very strong and 1 (16.6%) isolate had a mild enzymatic activity, while 1 (16.6%) isolate was proteinase negative. A statistically significant difference was observed between the two species regarding proteinase activity (P < 0.001).
5. Discussion
The C. parapsilosis complex known as the third most common important cause of candidiasis could easily colonize in hospital environments and healthcare workers. For this reason, discrimination of the C. parapsilosis complex to the species level and evaluation of their virulence factors are important in treatment and epidemiological studies. Various methods have been introduced for the identification of these species. Rycovska et al. reported the existence of these distinct species by analyzing the linear and circular forms of the mitochondrial genome and levels of heterozygosity in C. parapsilosis (4). Kosa et al. investigated mitochondrial genome size of these species and found that C. parapsilosis had bigger genome size than the other two species. They reported that these differences are due to the number of introns in the genome (14).
There are many studies regarding the molecular identification of the C. parapsilosis species complex by enzymes digestion of different genes amplified in PCR assay (1, 15, 16). Mirhendi et al. used the NlaIII enzyme for the digestion of SADH gene (1), Paula-Mattiello et al. (17) employed FKS1 gene and EcoRI enzyme, and another study (12) performed sequence analysis of the ITS region to discriminate this complex. Feng et al. used exon-primed intron-crossing (EPIC)-PCR assay for their identification (18). Asadzadeh carried out a study on clinical samples of Kuwaitian patients; he used multiplex PCR assay to present a low-cost detection method (19). Hass et al. applied the matrix assisted laser desorption ionization-time of flight mass spectrometry method for the discrimination of these species (20). Moreover, Barbedo et al. reported agreement in the results of four PCR-based methods (i.e., species-specific uniplex PCR, sequencing of the D1/D2 region of the LSU 28S rDNA gene, microsatellite typing, and PCR-RFLP of ITS) for species identification among 98 clinical isolates (5).
In this study, in accordance with the technique proposed by Neji et al. and Asadzadeh et al., we used Ban1 restriction enzyme for the analysis of SADH gene (16, 19). Using this method, we could identify C. parapsilosis sensu stricto (91.5%) as the major species among the group complex. The results of this study are in line with those of similar studies (16, 21-23). Although in many studies C. parapsilosis sensu stricto was the predominant species in this complex, there were differences in frequencies between the other two species. For example, the frequency of C. metapsilosis species was more than C. orthopsilosis in studies conducted by Neji et al. and Wu et al. (16, 23), while Borghi et al. (21) reported that C. orthopsilosis had higher frequency than the other species in their study.
We could not identify any species of C. metapsilosis. This result is in agreement with some reports from Iran and Denmark (1, 24), but it was not consistent with many reports from countries such as Taiwan, Italy, China, Mexico, and Brazil (21-23, 25, 26). Ge et al. reported high prevalence of C. metapsilosis in Chinese skin samples (22). In addition, many species of C. parapsilosis could not be identified when using BanI restriction enzyme because in some species there is no restriction site for this enzyme (1). Accordingly, care must be taken when using this enzyme for differentiation.
Although the contribution of proteinase virulence factor in C. parapsilosis remains nebulous, the role of aspartyl proteinases in the pathogenicity of C. albicans has been investigated during the past decades (13). There are ample studies on the evaluation of proteinase activity among the C.parapsilosis species complex. Paula-Mattiello et al. and Ge et al. reported that 93.1% and 80% of C. parapsilosis sensu stricto species were proteinase positive, respectively (8, 17). These data were almost similar to our finding, which was 70.76%. A high level of proteinase activity in C. orthopsilosis species was reported by Neji et al. (16); this result is in line with our data (i.e., 83.2%), but in the study conducted by Trevino-Rangel, this rate was lower (25). Overall, secretion of proteinase in the C. parapsilosis species complex could be considered as a virulence factor in the pathogenicity of these species. However, regarding C. metapsilosis further studies are required to arrive at this conclusion.
5.1. Conclusions
In the present work, we used the boiling method for DNA extraction, which is a prompt and efficient technique. Using PCR-RFLP with restriction enzyme Ban1 is a quick way to identify the species. In this study, we did not identify C. metapsilosis perhaps due to the limited number of samples. However, we observed a high level of proteinase activity in the identified species, and the level of activity in C. orthopsilosis was higher than that of C. parapsilosis species.