1. Background
Streptococcus agalactiae, or group B Streptococcus (GBS), is a Gram positive coccus with global distribution (1, 2). Streptococcus agalactiae cause neonatal meningitis in humans, mastitis in cattle, acute sepsis in rabbits, and meningoencephalitis as well as septicemia in fish (2-4). The capsular polysaccharide is contributing to the pathogenicity of S. agalactiae and it is useful in bacterial genotyping (5). Ten distinct capsular polysaccharide serotypes have been reported (Ia, Ib, II - IX) (6). Serotypes Ia and III are believed to be the most common and have the capability of infecting multiple host species (7, 8).
Serotyping alone may be insufficient to fully characterize isolates from different host species. Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) can provide a more robust analysis to investigate the genetic diversity of pathogen isolates (9-11). The MLST technique classifies the bacterial sequence types based on approximately 500 bp of seven housekeeping genes (12), which is due to the fact that they are varied over an evolutionary time scale (13). In contrast, PFGE discriminates among sequence types by analyzing diversity of genomic DNA digested with Sma I restriction enzyme (14).
Recent studies suggest that fish, human, and bovine S. agalactiae isolates are similar and can have a high degree of cross infectivity (1, 15). However, some reports suggest high diversity of S. agalactiae and there are distinct populations of human and bovine S. agalactiae isolates 21 (2, 16). To date, no studies have reported the genetic relatedness of S. agalactiae isolates among host populations in China. Therefore, we used serotyping, MLST, and PFGE methods to investigate the genetic diversity of S. agalactiae isolated from human, bovine, rabbit, and fish in China. These data are essential to understand the epidemiology and risk of S. agalactiae in China and elsewhere.
2. Objectives
In this study, it was aimed at determining the serotype diversity and molecular typing population structure of S. agalactiae isolated from human, bovine, rabbit, and fish by capsular polysaccharide serotyping, multilocus sequence typing, and pulsed field gel electrophoresis.
3. Methods
3.1. Ethics Statement
The entire experimental procedure was approved by the Committee of the Ethics on Animal Care and Experiments at Sichuan Agricultural University and was carried out in accordance with the approved guidelines.
3.2. Isolates Collection and Identification
A total of 56 strains identified as S. agalactiae from our lab (Center of Fish Disease at Sichuan Agricultural University, China) were used in this study. These samples were isolated from human (n = 16), bovine (n = 12), rabbit (n = 2), and fish (Schizothorax prenanti n = 6, Tilapian = 10, Schizo pygopsispylzovi n = 10). Streptococcus agalactiae ATCC51487 was used as a control. Fish isolates were grown at 28°C over 48 h in brain heart solid medium (OXOID, CM1135, Basingstoke, Hampshire, England), while human, bovine, and rabbit isolates were grown at 37°C for 24 h in brain heart solid medium. All strains were confirmed by Gram stain, morphology, and 16S rRNA sequencing. We identified the cfb gene, which encodes the CAMP factor for S. agalactiae, by polymerase chain reaction (PCR) and sequencing: Forward primer (5′-AGCTTAGTTATCCCA AATCCCAT-3′) and reverse primer (5′-GTGCCAACCCTGAGACAGTT-3′) (17).
3.3. Identification of Capsular Types
Genomic DNA was extracted from each isolate by TIANamp bacteria DNA kit (TIANGEN Biotech Inc. Beijing, China). Collected DNA was washed and dissolved in 25 µL of buffer TE and stored at 4°C. The capsular polysaccharide serotypes were identified by multiplex polymerase chain reaction using the method of Imperi (6).
3.4. MLST Analysis
MLST was performed based on seven housekeeping genes, primers are listed in Table 1 (18). The 25 µL PCR reaction system was: 12.5 µL 2 × Taq PCR Master Mix, 1.0 µL of each primer, 2 µL DNA template, and 8.5 µL distilled water. The PCR amplification procedure was: 5 min at 95°C, 33 cycles at 95°C, 55°C, 72°C for 45 s, and one cycle at 72°C for 10 min. The PCR products were sequenced using the primers listed in the Table 1 and the sequences were submitted to the S. agalactiae MLST database to determine the allele numbers and sequence types; the new sequence types (STs) were submitted to PubMLST website. The eBURST algorithm (19) was performed to infer the hypothetical phylogenetic relationship among STs.
| Primer Name | Primer Sequence (5´ - 3´) | Fragment Length (bp) |
|---|---|---|
| adhP | 498 | |
| F | GGTGTGTGCCATACTGATTT | |
| R | ACAGCAGTCACAACCACTCC | |
| pheS | 501 | |
| F | ATATCAACTCAAGAAAAGCT | |
| R | TGATGGAATTGAATGGCTATG | |
| atr | 501 | |
| F | CGATTCTCTCAGGCTTTGTTA | |
| R | AAGAAATCTCTTGTGCGGAT | |
| glnA | 498 | |
| F | AATAAAGCAATGTTTGATGG | |
| R | GCATTGTTCCCTTCATTATC | |
| sdhA | 519 | |
| F | AACATAGCAGAGCTCATGAT | |
| R | GGGACTTCAACTAAACCTGC | |
| glcK | 459 | |
| F | GGTATCTTGACGCTTGAGGG | |
| R | ATCGCTGCTTTAATGGCAGA | |
| tkt | 480 | |
| F | ACACTTCATGGTGATGGTTG | |
| R | TGACCTAGGTCATGAGCTTT |
The Relevant Experimental Parameters of the Housekeeping Gene Locus of Streptococcus agalactiae.
3.5. PFGE Analysis
The PFGE was performed according to the method described by Chen (20), with slight modifications. Major steps included: S. agalactiae cells cultured in brain heart infusion broth for a mid-logarithmic phase were harvested and resuspended in solution buffer (100 mM Tris-HCl, 100 mM EDTA, PH 7.6); S. agalactiae cells were digested with 10 mg/mL lysozyme, 20 mg/mL proteinase K, and 1mg/mL mutanolysin (TaKaRa Biotechnology Co. Ltd. Dalian, China); mix up the bacterial suspension of equal parts 1.5% melting agarose (Bio-Rad Laboratories. Beijing, China); genomic DNA was digested with 50 U of restriction endonuclease Sma I (Thermo Fisher Scientific. Chengdu, China) at 37°C for 30 min; electrophoresis was performed in CHEF Mapper® XA (Bio-Rad Laboratories. Beijing, China) with a pulse ramping between 10 - 20s at 14°C for 18 h and 6 V/cm. The PFGE results were analyzed using Quantity One v.4.62 (Bio-Rad Laboratories. Beijing, China) and a dendrogram was created from a matrix of band matching using the unweighted pair group method with the arithmetic means analysis. PFGE cluster was made based on a threshold of 77% similarity.
4. Results
4.1. Serotype Identification
All isolates were identified as S. agalactiae, according to species-specific PCR. Our 56 S. agalactiae isolates belonged to two serotypes (Ia and III); serotype Ia was the most common (85.7%; fish n = 22, human n = 12, bovine n = 12, rabbit n = 2). Fish and human isolates possessed of both serotypes, while bovine and rabbit isolates solely belonged to serotype Ia.
4.2. MLST Typing of S. agalactiae
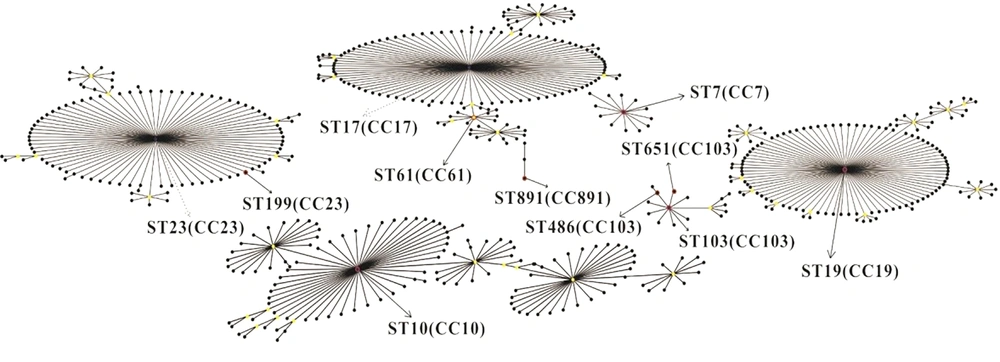
The MLST revealed a total of nine STs among 56 isolates (Table 2), with seven corresponding to a single host. The remaining two STs (ST19, ST103) belonged to two hosts. ST891 was found in the largest number of isolates (n = 20, 35.7%), followed by ST10 (n = 8, 14.3%) and ST103 (n = 8, 14.3%). Additionally, ST19 and ST486 were found in six isolates (10.7%). The rest of STs were found in two isolates (3.6%). We discovered a new ST (ST-891) and entered it in S. agalactiae MLST database. In general, the STs had no obvious correlation with source of the hosts. The same STs were identified in multiple hosts (e.g., ST103 in fish n = 2 and cattle n = 6), and the same host could have a variety of STs (e.g., human: ST10, ST61, ST199, ST651). In addition, the eBRUST algorithm results showed that all isolates were grouped in seven clonal complexes: CC7, CC10, CC17, CC19, CC23, CC103, and CC891 (Figure 1), among which ST61 was a subgroup of CC17. Both ST486 and ST651 were a single-locus variant (SLV) for ST103. ST199 was SLV for ST23. Lastly, ST7, ST10, ST19, ST103, and ST891 were the founders of clonal complexes CC7, CC10, CC19, CC103, and CC891, respectively.
| Typing | Allele Number | Number of Isolates(n) | ||||||
|---|---|---|---|---|---|---|---|---|
| adhP | pheS | atr | glnA | adhA | glcK | tkt | ||
| ST7 | 10 | 1 | 2 | 1 | 3 | 2 | 2 | 2 (rabbit) |
| ST10 | 9 | 1 | 4 | 1 | 3 | 3 | 2 | 8 (human) |
| ST19 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 6 (fish n = 4, human n=2) |
| ST61 | 13 | 1 | 1 | 13 | 1 | 1 | 1 | 2 (human) |
| ST103 | 16 | 1 | 6 | 2 | 9 | 9 | 2 | 8 (fish n = 2, bovine n = 6) |
| ST199 | 5 | 4 | 6 | 3 | 2 | 1 | 1 | 2 (human) |
| ST486 | 16 | 1 | 6 | 2 | 9 | 1 | 2 | 6 (bovine) |
| ST651 | 16 | 1 | 6 | 70 | 9 | 9 | 2 | 2 (human) |
| ST891 | 54 | 17 | 31 | 81 | 26 | 25 | 19 | 20 (fish) |
Correlation Between Allele Numbers, Sequence Types and Bacterial Sources of Streptococcus agalactiae.
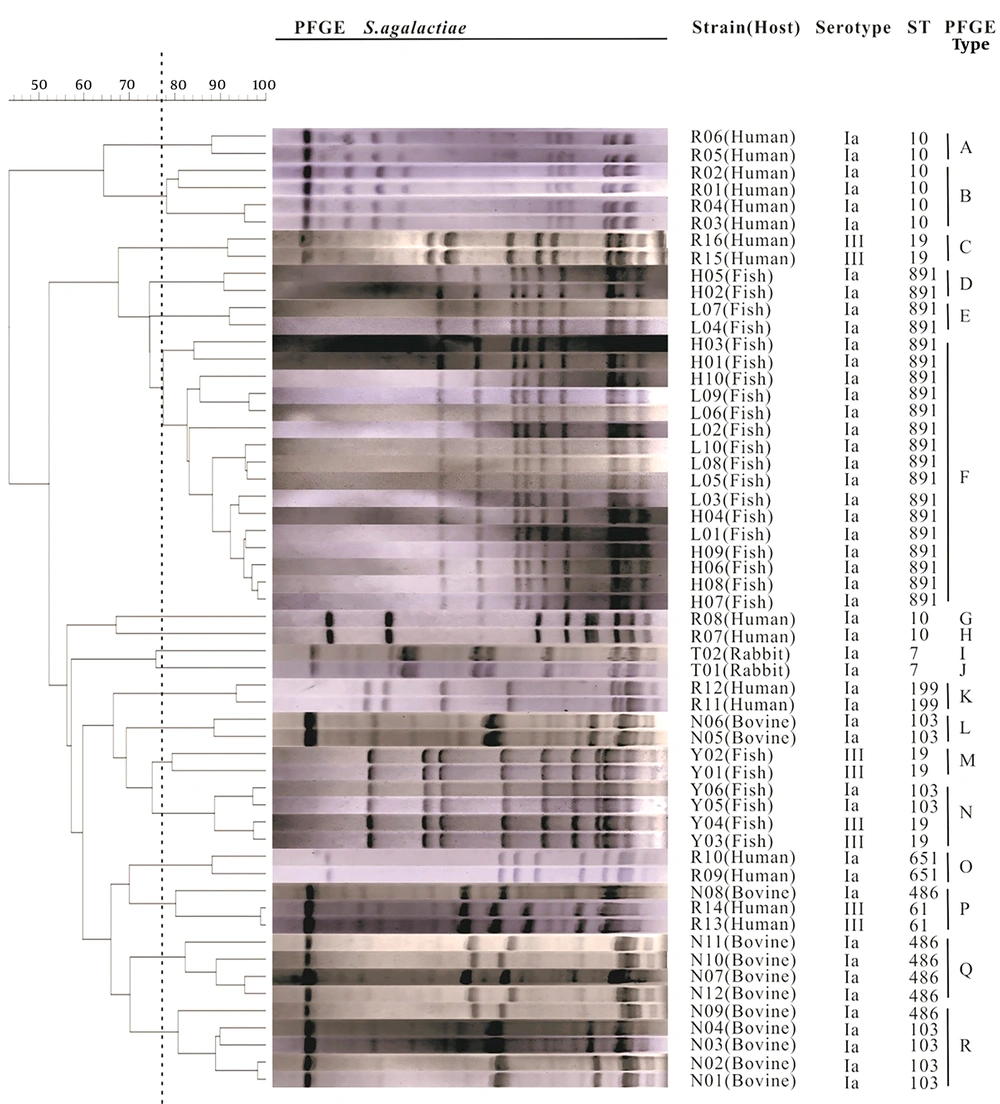
4.3. Diversity of PFGE Genotypes
The PFGE results indicated that a total of 18 different PFGE patterns were obtained in these isolates (Figure 2), among which four isolates showed an individual PFGE profile. Sixteen fish isolates belonging to ST891 showed an identical PFGE profile, which was the most prevalent. Meanwhile, the PFGE diversity of human isolates was even larger than that observed in other host isolates. The cluster P was found in human and bovine isolates. The rest of PFGE clusters contained one kind of isolate. Categorical analysis revealed that the PFGE clusters were independently distributed in single source of isolate, except cluster P.
Cluster analysis was performed with quantity one software (Bio-Rad Laboratories) by using the UPGMA method. The 56 S. agalactiae strains are divided in 18 clusters as defined by the vertical dotted line. The serotype and ST data are plotted next to the dendrogram.
5. Discussion
Comparative analysis of the molecular characteristics of S. agalactiae strains isolated from different hosts was investigated (1, 15). However, the genotyping diversity of S. agalactiae isolated from different host species in China was unclear. In this study, two serotypes were identified, including Ia (n = 48, 85.7%) and III (n = 8, 14.3%). This result corresponds with some reports that serotypes Ia and III are the predominant serotypes of S. agalactiae (21, 22), and illustrates the relatively simple serotype population structure in China. Bovine and rabbit isolates belonged to serotype Ia, while human and fish isolates were mostly serotype Ia with a few cases of serotype III.
MLST analysis showed that a total of nine STs and seven CCs were identified. Among them, a novel STs (derived from fish) was found and submitted to the S. agalactiae MLST database. This ST (ST891) was the most common (n = 20, 35.7%) among isolates. Isolates from different hosts could be classified into different clonal complexes, except CC103, which possessed three STs (ST103, ST486, ST651) detected in human, bovine, and fish isolates. Sixteen human isolates had several sequence types including ST19, ST61, ST10, ST199, and ST651. In contrast, the two rabbit isolates belonged to ST7, which was the first time to be confirmed. A total of 18 PFGE types were identified in this study. Each cluster was associated with a single isolate source, except the cluster P, which contained human and bovine isolates. However, isolates from different hosts still presented quite dramatic PFGE typing diversity. The PFGE result in this study is in line with a previous observation (23) where S. agalactiae isolated from different hosts exhibit high genetic diversity and few relationships.
Additionally, the serotypes had no obvious correlation with STs and PFGE types. The genetic analysis showed that isolates of serotype Ia were distributed in different STs and PFGE types. Serotype III included both ST19 and ST61, which was also grouped in four clusters (C, M, N, P). Furthermore, the same ST/CPS type could be grouped in different PFGE patterns. For example, ST103/Ia contained isolates from bovine (n = 6) and fish (n = 2), and was grouped in two different PFGE clusters (L and R). In addition, the ST19/III contained isolates from human (n = 2) and fish (n = 4) and were grouped in cluster C and N. Therefore, the bovine-fish isolates of ST103/Ia and the human-fish isolates of ST19/III may not be able to infect each other. This finding contradicts previous research suggesting that serotype III in human strains can infect fish (10). Additional research is needed for investigating the possibility of S. agalactiae isolated from human, rabbit, bovine, and fish cross infecting. If cross-infection is possible, future research should attempt to identify the responsible genes, which could lead to gene-editing therapies that limit zoonotic transmission.
5.1. Conclusion
Our results showed that serotypes, STs, and PFGE profiles had no obvious correlation with 56 isolates from a range of different hosts. The results also revealed that serotype Ia was the preponderant type of S. agalactiae isolated from four kinds of host species in China. The genetic diversity and clonal population structure were evaluated by MLST and PFGE. Nine ST and seven CC were obtained, among which ST651 from the human was a single-locus variant for ST103 from cattle, this suggested more possibilities for cross infection at genetic level. In the future, cross-infection possibility for S. agalactiae strains isolated from human, rabbit, cattle, and fish will be confirmed.