1. Background
Acinetobacter baumannii are non-motile Gram-negative bacteria, which can be found widespread in soil, water, and various samples of humans or animals (1, 2). These bacteria are a leading cause of endocarditis, skin and soft tissue infections, meningitis, urinary tract infections, and pneumonia (3, 4), partly due to resistant features of this opportunistic microbe to a variety of antibiotics, resulting in rapid emergence of multidrug and extremely drug resistant strains (5). This is the reason these bacteria have become one the most successful pathogens in the modern healthcare system and endemic in hospitals, especially in patients at the intensive care unit (ICU) (6). Additionally, A. baumannii can survive in both dry and damp hospital environmental conditions, even for lengthened periods of time (1, 7). Therefore, development of non-antibiotic-based intervention strategies seems urgent (8). A variety of risk factors have been proposed in acquiring A. baumannii infection. These include immunosuppression, unscheduled hospital admission, respiratory failure at ICU admission, previous antimicrobial therapy, previous sepsis in the ICU, and the invasive procedures index (9).
Increase of the number of patients with acquired nosocomial Acinetobacter infectious outside the ICU has been recently reported, which is alarming and shows the ability of this pathogen to become a major healthcare barrier (10). These findings suggest urgent need for the development of novel strategies to fight this multidrug-resistant pathogen (11). Vaccines are one of the most effective medical plans for the control of infections worldwide and are usually designed based on weekend bacteria or their purified immunogenic parts, such as proteins or polysaccharide wraps. However, such vaccines are not very effective for inducing cell-mediated immunity against intracellular pathogens (12, 13). Recently, a new strategy has been proposed, which is based on the production of recombinant protein from infectious agents with acceptable antigenic potential. In this approach, which is also known as second generation of vaccines, a gene encoding antigenic protein can be cloned in an expression plasmid, expressed in a host cell, including bacteria or yeast.
The recombinant protein was then purified to be tested as a vaccine (13). Such strategy has been recently proposed and applied to develop hepatitis B virus (HBV), human papillomavirus (HPV), human immunodeficiency virus (HIV), tuberculosis (TB), and malaria vaccine (14). Besides, clinical trials have shown that these vaccines are safe and well-tolerated (15). In Gram-negative bacteria, the outer membrane is a safeguard layer against external perilous agents, such as foreign enzymes and detergents (16). It has been documented that outer membrane proteins (OMPs), such as OmpA, promote the virulence features of A. baumannii through their critical role in biofilm formation or bacterial adhesion on epithelial cells (2, 17). These proteins can also cause bacteria to withstand against environmental stress (18). In a reverse vaccinology study by Moriel, the ompA gene was introduced as one the potent genes to develop DNA vaccine against A. baumannii (19).
2. Objectives
It has been indicated that OmpA are a potent inducer of native and adaptive immune responses in vivo (20). Notably, 2% to 3% of the Gram negative bacteria genome consists of genes encoding the OMPs. Therefore, these proteins are recently proposed as proper candidates for recombinant vaccine development (21). Here, the ompA gene of A. baumannii was extracted from antibiotic resistant A. baumannii-infected clinical samples and efficiently cloned and expressed in pET32a (+) expression system, one of the most effective systems for gene expression and production of recombinant proteins (22).
3. Methods
3.1. Ethics Statement
This study (experimental and cross-sectional) was part of a Ph.D. thesis, which was ethically approved by the Islamic Azad University of Marvdasht, Iran (IR.MIAU.REC.1397.802), and experimentally performed at Islamic Azad University of Shahrekord, Iran.
3.2. Bacterial Isolates
Sixty-seven samples were collected from different clinical cases of pneumonia (sputum, n = 25 isolates), wound pus (n = 18 isolates), septicemia (blood, n = 14 isolates), and urinary tract infections (n = 10 isolates) at the ICU and different wards of Imam Ali and Kashani hospitals of Shahrekord, southwest Iran, during September 2015 to April 2016. To avoid multiple entries from a single patient, only the first sample for a given patient was included. All samples were cultured on solid media (MacConkey; Merck, Germany and blood agars). The A. baumannii was diagnosed at a microscopic scale as well as biochemical and bacteriological standard tests. The two later assays included growth at 44°C, Gram staining, MacConkey agar, Sulfur Indole Motility media (SIM) assay, oxidase-catalase assay, hemolysis test, triple sugar iron (TSI) test, arginine hydrolysis assay, ornithine decarboxylase test, citrate assay, and gelatin hydrolysis. The isolates were stored at -70°C in trypticase soy broth (Merck, Germany), supplemented with 15% glycerol, before use.
3.3. Antimicrobial Susceptibility Tests
The disk diffusion method was used to examine the susceptibility of all isolates to 17 antimicrobial agents, as recommended and outlined by the National Committee for Clinical Laboratory Standards (CLSI, 26th edition 2016, USA) (23). Antimicrobial agents, including tobramycin (10 μg), tetracycline (30 μg), oxacillin (1 μg), trimethoprim (5 μg), gentamicin (10 μg), meropenem (10 μg), imipenem (10 μg), colistin (10 μg), ceftriaxone (30 μg), ampicillin-sulbactam (10/10 μg), co-trimoxazole (30 μg), ciprofloxacin (5 μg), cefepime (30 μg), ceftazidime (30 μg), aztreonam (30 μg), amikacin (30 μg), and piperacillin-tazobactam (100/10 μg) were used in this study. All of the antibiotics were purchased from MAST, UK or Liofilichem, Italy. The isolates were grown on trypticase soy agar plates, supplemented with 5% sheep blood at 37°C overnight. The A. baumannii ATCC 19606 and Escherichia coli ATTC 25922 were purchased from the bacterial repository of Iranian Biological Resource Center (IBRC, Karaj, Iran) and considered for quality control purposes (positive and negative control) in the antibacterial susceptibility assay.
3.4. Detection of MDR, PDR and XDR Isolates
Defining of multidrug resistance (MDR), pan drug resistant (PDR), and extreme drug resistance (XDR) in A. baumannii isolates was done according to new standardized international document (24), with the results of antimicrobial susceptibility of A. baumannii to all antimicrobial agents listed in Table 1. Briefly, MDR is defined as non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories, XDR was described as non- susceptibility to ≥ 1 agent in ≤ 2 antimicrobial categories, and PDR was defined as non- susceptibility to all antimicrobial agents listed.
| Antibiotic | S | I | R |
|---|---|---|---|
| Penicillins | |||
| Ampicillin-Sulbactam (SAM) | 16 (53.34) | 4 (13.33) | 10 (33.33) |
| Piperacillin plus Tazobactam (TZP) | 7 (23.33) | 4 (13.33) | 19 (63.34) |
| Oxacillin (OXA) | 4 (13.33) | 9 (30) | 17 (56.67) |
| Cephalosporins | |||
| Ceftazidime (CAZ) | 1 (3.33) | 2 (6.67) | 27 (90) |
| Ceftriaxone (CRO) | 1 (3.33) | 4 (13.33) | 25 (83.34) |
| Cefepime (CPM) | 0 (0) | 2 (6.67) | 28 (93.33) |
| Carbapenems and β-lactams | |||
| Imipenem (IPM) | 10 (33.33) | 3 (10) | 17 (56.67) |
| Meropenem (MEM) | 5 (16.67) | 9 (30) | 16 (53.33) |
| Aztreonam (ATM) | 4 (13.33) | 3 (10) | 23 (76.67) |
| Tetracyclines | |||
| Tetracycline (TET) | 9 (30) | 6 (20) | 15 (50) |
| Aminoglycosides | |||
| Amikacin (AMK) | 5 (16.67) | 4 (13.33) | 21 (70) |
| Tobramycin (TOB) | 4 (13.33) | 8 (26.67) | 18 (60) |
| Gentamicin (GEN) | 7 (23.23) | 3 (10) | 20 (66.67) |
| Sulphonamides | |||
| Co-trimoxazole (TS) | 4 (13.33) | 9 (30) | 17 (56.67) |
| Folate pathway inhibitor | |||
| Trimethoprim (TP) | 5 (16.67) | 12 (40) | 13 (43.33) |
| Quinolones | |||
| Ciprofloxacin (CIP) | 3 (10) | 7 (23.33) | 20 (66.67) |
| Lipopeptides | |||
| Colistin (CO) | 23 (76.67) | 4 (13.33) | 5 (10) |
aValues are expressed as number (percentage).
3.5. Extraction of Genomic DNA from Acinetobacter baumannii
The first step to development of the ompA-containing construct was the extraction of genomic DNA from resistant isolates, using the DNA extraction kit (Qiagen DNA Mini Kit, Iran), according to manufacturer’s instructions. The integrity of extracted DNA was evaluated through 1% agarose gel electrophoresis. The purity and concentration of DNA samples was calculated by Nano-Drop ND-1000 (PeqLab Ltd., UK) at wavelengths of 230, 260, and 280 nm.
3.6. Polymerase Chain Reaction (PCR)
The ompA gene (accession number: AY485227) coding sequence was amplified using specific primers, as the 1112 bp product. The primer sequences were designed using Gene Runner software version 5. The sequences of primers was as follow: Forward: 5’-AGGAAGCTTATGAAATTGAGTCGTATTGC-3’ and reverse: 5’-CGTCTCGAGTTTTTACTGTTCAAGAACTC-3’. A restriction site for HindIII and XhoI were included in forward and reverse primers, respectively. The PCR reaction was optimized in a total volume of 25 μL, containing 2.5 μL 10X PCR buffer, 2 mM MgCl2, 100 mM dNTPs, 1 mM of each primer, 1 μg DNA sample, and 1 unit of Taq DNA polymerase (Fermentas, Germany). The PCR was programed as 35 cycles of initial denaturation at 95°C for five minutes; denaturation at 94°C for one minute, annealing at 61°C for one minute, extension 72°C for one minute, and final extension at 72°C for five minutes. The PCR products were checked on 2% agarose gel electrophoresis and bands were visualized under UV trans-illuminator (Uvtec, UK).
3.7. Cloning ompA Gene
The ompA amplicon were purified from the gel, using the gel extraction kit (Bioneer, Korea), according to the manufacturer’s instructions and used for cloning in T/A cloning vector pTZ57RT (Thermo Scientific, Lithuania). The recombinant construct was then subjected to transform E. coli NovaBlue competent cells, using 42°C heat shock and calcium chloride method. Transformed competent cells were then cultured in LB agar media (Merck Co., Germany), containing 20 mg/mL X-gal, 0.1 M IPTG, and 100 μg/mL ampicillin at 37°C overnight. The white colonies were re-cultured in LB broth media, supplemented with 100 μg/mL ampicillin at 37°C overnight. The recombinant vector was extracted from bacterial cells using Plasmid Mini Extraction Kit (Bioneer, Korea), according to the user manual. The HindIII/XhoI double digestion was performed to confirm whether ompA has been correctly inserted in the vector. Polymerase chain reaction-sequencing was also applied to validate the accurate cloning.
3.8. Sub-Cloning of ompA Gene in the Expression Vector pET-32a (+)
Using HindIII/XhoI double digestion, the 1112 bp corresponding to ompA amplicon was removed from the T/A recombinant pTZ57RT vector and considered to ligate with HindIII/XhoI linearized pET-32a. The T4 ligase enzyme was used to generate the pET-32a-ompA recombinant expression vector. The E. coli NovaBlue competent cells were used as a transformation host, which were cultured in LB agar media, containing 100 μg/mL of ampicillin. The pET-32a-ompA recombinant construct was validated by PCR-sequencing, using ompA-F and ompA-R primers, as well as HindIII/XhoI double digestion.
3.9. SDS-PAGE Analysis
The pET32a-ompA was introduced in the E. coli Rosetta-gami™ 2 (DE3), using the CaCl2 method. Transformed cells were then cultured on the LB agar, containing 100 μg/mL ampicillin and 35 μg/mL chloramphenicol. The recombinant E. coli DE3 was grown in 5 mL LB broth, containing ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL) on a shaking incubator overnight at 37°C. Then, 500 mL of culture was inoculated in 50 mL of LB broth medium at 37°C with agitation at 200 rpm. The cells were grown until the optical density reached 0.6 to 0.8 at 600 nm. Expression of the OmpA protein was induced by the 0.9 mM IPTG and incubated for five hours at 250 rpm/min, at 37°C. The cells were harvested by centrifugation at 9000 rpm. The harvested cells then subjected for sonication; Ten short bursts of 10 seconds followed by intervals of 30 seconds on ice to obtain the viscous cell lysate. Whole cell lysate was then analyzed on 12% SDS-PAGE. The IPTG-induced E. coli with empty pET32a was also considered as the negative control. The mentioned control confirms that the empty vector does not express any proteins with similar molecular weight to the target protein (OmpA).
3.10. RNA Extraction
Total RNA was extracted from 107 recombinant E. coli DE3 containing pET32a-ompA, induced by IPTG, using RNX-Plus reagent (SinaClon, Iran), according to the user manual. The concentration and purity of the genomic DNA were evaluated by Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), reading at wave length 230 nm, 260 nm, and 280 nm. The RNA was considered pure if the absorbance ratio of 260 nm/280 nm and 260 nm/230 nm was around two. The integrity of extracted RNAs was also evaluated on 1% agarose gel electrophoresis.
3.11. Complementary DNA (cDNA) Synthesis and RT-PCR
A reaction containing 500 ng/µL RNA, 100 mM dNTPs, 10 µL 5x RT-PCR buffer, 200 mM oligdT primer, and 1 U/µL M-MuLV reverse transcriptase, (Invitrogen, USA) in a total volume of 20 μL was prepared for cDNA synthesis. The mixture was incubated at 42°C for one hour, followed by 15 minutes at 70°C for enzyme inactivation. The ompA transcript was then amplified using the specific primers, as mentioned above. The reaction was performed in total volume of 20 μL, containing 50 mM MgCl2, 0.25 mM of each dNTP, 2 pmol/L of each primer, 400 ng/ µL cDNA templates, and 1 U of Taq DNA polymerase (Fermentas, Germany). The final volume was adjusted with nuclease-free water. The PCR was programmed in a thermocycler as five minutes of denaturation at 95°C for initial denaturation, followed by 25 cycles of denaturation; 94°C for one minute, annealing; 61°C for one minute, extension; 72°C for 45 seconds, final extension; 72°C for two minutes. A sample without DNA template (NTC) was included in all PCR reactions as the negative control. The PCR products were then analyzed on 1% agarose gel electrophoresis, dyed with 2% ethidium bromide, and product bands were visualized under ultraviolet light.
3.12. Statistical Analysis
All the data was entered in the SPSS version 22 (SPSS Inc., Chicago, IL) software for descriptive analysis. A two-tailed P < 0.05 was considered statistically significant. In susceptibility assay, data were classified as susceptible and non-susceptible (including sensitive; S, intermediate; I and resistant groups; R).
4. Results
4.1. Antibiotic Resistance Pattern of Acinetobacter baumannii
Out of 67 collected samples, 30 samples (44.48%) were diagnosed as A. baumannii positive. These bacteria were recovered from sputum (n = 15 isolates), wound pus (n = 7 isolates), blood (n = 6 isolates), and urine (n = 2 isolates). These 30 isolates were subjected to antibiotic susceptibility assay, the data of which has been summarized in Table 1. This study found that these 30 bacterial isolates were highly resistant to all antibiotics, except colistin and ampicillin-sulbactam. In this way, 66.7% to 93.3% of the strains were resistant to the antibiotics cefepime, ceftazidime, ceftriaxone, aztreonam, amikacin, gentamicin, and ciprofloxacin. The distribution of resistant isolates in different samples is summarized in Table 2. As illustrated in Table 3, 29 isolates were MDR and one isolate was resistant to all categories of tested antibiotics, which places it in the PDR group.
| Source | SAM | AMK | ATM | CAZ | CPM | CIP | TS | CRO | CO | TOB | IPM | MEM | GEN | TP | OXA | TET | TZP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Septum (n = 25) | |||||||||||||||||
| S | 3 | 1 | 0 | 1 | 0 | 1 | 2 | 1 | 18 | 1 | 4 | 1 | 3 | 2 | 1 | 2 | 3 |
| I | 1 | 0 | 0 | 1 | 0 | 2 | 3 | 0 | 2 | 2 | 1 | 4 | 0 | 5 | 4 | 1 | 1 |
| R | 0 | 2 | 2 | 2 | 6 | 1 | 1 | 10 | 1 | 4 | 1 | 2 | 7 | 5 | 2 | 0 | 13 |
| Wound pus (n = 18) | |||||||||||||||||
| S | 3 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 3 | 1 | 3 | 2 | 2 | 1 | 1 | 3 | 1 |
| I | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 4 | 1 | 5 | 1 | 3 | 2 | 3 | 1 |
| R | 1 | 2 | 4 | 8 | 5 | 3 | 3 | 2 | 2 | 6 | 6 | 3 | 5 | 6 | 3 | 0 | 4 |
| Septicemia blood (n = 14) | |||||||||||||||||
| S | 5 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 3 | 1 |
| I | 1 | 0 | 1 | 0 | 1 | 3 | 2 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| R | 4 | 4 | 8 | 8 | 6 | 10 | 10 | 6 | 2 | 0 | 6 | 3 | 5 | 5 | 2 | 7 | 2 |
| Urinary tract infection (n = 10) | |||||||||||||||||
| S | 5 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | 2 |
| I | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 0 | 0 | 1 | 3 | 1 | 2 | 1 |
| R | 5 | 13 | 9 | 9 | 11 | 6 | 3 | 7 | 0 | 8 | 5 | 8 | 3 | 4 | 10 | 8 | 0 |
aData has been presented as the number of isolates in each group.
| Antibiotics Resistant Patterns | Number of Antibiotics in the Resistant Pattern | Sample No. | Frequency in 30 Isolates | Type of Resistant Pattern |
|---|---|---|---|---|
| CPM, CAZ, CRO, ATM, AMK, CIP, TZP, TOB, TS, IPM, OXA, MEM, TET, SAM | 14 | 1 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, GEN, CIP, TZP, TOB, TS, IPM, OXA, MEM, TET, TP, SAM, CO | 17 | 2 | 1 | PDR |
| CPM, CAZ, CRO, ATM, AMK, GEN, CIP, TZP, TOB, TS, OXA, MEM, TET, TP | 14 | 3 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, CIP, TZP, OXA, TET, TP | 10 | 4 | 1 | MDR |
| CPM, CAZ, GEN, TZP, TOB, TS, IPM, TP | 8 | 5 | 1 | MDR |
| CPM, CAZ, CRO, AMK, GEN, TZP, TS, OXA, SAM | 9 | 6 | 1 | MDR |
| CPM, CRO, ATM, AMK, CIP, TZP, MEM | 7 | 7 | 1 | MDR |
| CPM, CAZ, CRO, ATM, GEN, TZP, TOB, IPM, OXA, TET, SAM | 11 | 8 | 1 | MDR |
| CPM, CAZ, CRO, ATM, TZP, TOB, TS, IPM, TET, TP | 10 | 9 | 1 | MDR |
| CPM, CAZ, ATM, AMK, CIP, TOB, IPM, SAM | 8 | 10 | 1 | MDR |
| CPM, CAZ, CRO, CIP, TS, IPM, OXA, TP, SAM | 9 | 11 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, GEN, TZP, IPM, MEM, CO | 10 | 12 | 1 | MDR |
| CPM, CRO, ATM, AMK, GEN, CIP, TOB, TS, OXA, TET | 10 | 13 | 1 | MDR |
| CPM, CAZ, CRO, CIP, TZP, TS, TP, SAM | 8 | 14 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, TZP, IPM, OXA, MEM, SAM | 10 | 15 | 1 | MDR |
| CPM, CAZ, CRO, AMK, GEN, CIP, TS, OXA | 8 | 16 | 1 | MDR |
| CPM, CAZ, ATM, AMK, GEN, TOB, TS, MEM, TET | 9 | 17 | 1 | MDR |
| CPM, CAZ, CRO, AMK, GEN, TZP, TOB, IPM, MEM, TP, SAM, CO | 12 | 18 | 1 | MDR |
| CAZ, CRO, ATM, GEN, CIP, TS, OXA, MEM | 8 | 19 | 1 | MDR |
| CPM, CAZ, AMK, GEN, CIP, TZP, TOB, IPM, MEM, TP | 10 | 20 | 1 | MDR |
| CPM, CAZ, ATM, AMK, GEN, CIP, TZP, TOB, TS, IPM, OXA, MEM | 12 | 21 | 1 | MDR |
| CPM, CRO, ATM, AMK, GEN, CIP, TOB, TS, OXA, MEM, TET | 11 | 22 | 1 | MDR |
| CPM, CAZ, CRO, ATM, GEN, CIP, TZP, IPM, MEM, SAM | 10 | 23 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, CIP, TS, IPM, MEM, TET, TP, SAM | 12 | 24 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, CIP, TZP, TOB, OXA, MEM, TP | 11 | 25 | 1 | MDR |
| CAZ, CRO, ATM, AMK, GEN, TS, OXA, TET, TP | 9 | 26 | 1 | MDR |
| CPM, CAZ, CRO, ATM, GEN, CIP, TOB, IPM, MEM, TET, TP | 11 | 27 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, GEN, TZP, TOB, IPM, TET | 10 | 28 | 1 | MDR |
| CPM, CAZ, CRO, ATM, AMK, GEN, CIP, TOB, TS, OXA, TET | 11 | 29 | 1 | MDR |
| CPM, CAZ, CRO, ATM, GEN, CIP, TZP, TOB, IPM, OXA, TET | 11 | 30 | 1 | MDR |
4.2. The Acinetobacter baumannii-Associated ompA Gene Was Successfully Cloned and Sub-Cloned inpTZ57RT and pET32a (+) Vectors
The ompA gene was amplified using PCR, as 1112 bp product (Figure 1). The amplicon was then subjected to cloned in the pTZ57RT vector. The PCR-DNA sequencing confirmed that the inserted ompA was completely aligned with the 98 recorded sequence of ompA for A. baumannii in gene bank. The degree of identity varied from 98% to 100%. The list of isolates, which had maximum sequence identity with the ompA sequence of the present study has been presented in Supplementary File. The obtained data from HindIII and XhoI double digestion also confirmed that ompA was correctly incorporated in the pTZ57RT vector. The presence of bands 1112 bp and 2886 bp during the electrophoresis was consistent with this fact. These bands belonged to the target gene and vector, respectively (Figure 2). Consequently, 1112-bp fragment of ompA was directly cloned at the HindIII and XhoI site of pET-32a, resulting in a recombinant plasmid, named pET-32a(+)-ompA. The recombinant plasmid (pET-32a(+)-ompA) was confirmed by PCR and HindIII/XhoI double digestion. The obtained data showed that the recombinant plasmid contained the ompA target gene. The bands 1112 bp and 5900 bp were observed during the electrophoresis, as expected. These bands were related to the target gene and vector, respectively (Figures 3A and 3B).
Cloning of ompA in pET32a; colony PCR of ompA. A, lane M, 1 Kb DNA marker; lane 1 - 7, an amplification of complete ompA gene (1112 bp) from pET32a-ompA; lane 8, negative control. B, analysis of recombinant expression vector (pET32a-ompA) digested by two restrictive endonucleases HindIII and XhoI. Lane 1 - 6, 5900 bp segment pET32a and fragment 1112 bp ompA; lane M, 1 Kb DNA marker; lane 7, pET32a undigested.
4.3. The rOmpA Protein Was Expressed in the E. coli Through Recombinant pET-32a(+)-ompA Construct
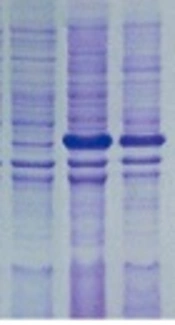
The pET-32a(+)-ompA construct was transferred to E. coli Rosetta-gami and expression of recombinant protein was evaluated through IPTG induction. As illustrated in Figure 4, the presence of a ~38 kDa protein band (as expected for OmpA protein) on SDS-PAGE showed that the pET32a vector was capable of expressing the OmpA protein.
SDS-PAGE analysis of A. baumannii OmpA. SDS-PAGE was used to analyze the expression of rOmpA. Recombinant vectors pET32a-ompA and pET32a (+) were transformed to E. coli Rosetta-gami™ 2 (DE3) and induced by 0.9 mM IPTG for 4 h at 37°C. All the samples were analyzed by SDS-PAGE, and the protein was stained with Coomassie Blue in the gel. Lane M, molecular mass marker in 10 kDa; lane 1, whole bacterial lysate of the E.coli Rosetta-gami containing empty pET32a; lane 2, non-induced E.coli Rosetta-gami including pET32a-ompA; lane 3 - 4, the whole bacterial lysate and supernatant of the E.coli Rosetta-gami™ 2 (DE3) contained recombinant pET32a-ompA which was induced; lane 5, Blank control.
4.4. The ompA-pET32a Was Successfully Expressed in Recombinant E. coli
The RNA was extracted from the recombinant E. coli, containing pET32a-ompA induced by IPTG. The cDNA was then synthesized from the transcripts to be amplified in RT-PCR, using specific primers of ompA. The presence of 1112 bp band confirmed that the pET32a vector can express the ompA coding sequence (Figure 5).
5. Discussion
Over the past two decades, A. baumannii was reported as one the major health problems due to the emergence of resistant strains, making it a dangerous opportunistic nosocomial pathogen (25). In a commentary published by the Center for Disease Control and Prevention during year 2004, A. baumannii was introduced as the leading cause of 80% of nosocomial infections and second pathogen isolated from hospitalized patients after Pseudomonas strains (26). Nosocomial septicity caused by the A. baumannii strains are progressively becoming resistant to a wide range of antibiotics (27). The current study revealed the resistant pattern of 30 strains of A. baumannii isolated from 67 clinical samples. All of the included patients were hospitalized at the ICU or different wards of Shahrekord hospitals. The samples were collected from sputum, urine, wound pus, and septicemia although most of them were obtained from sputum culture.
It was found that all of the 30 isolates of A. baumannii were resistant to examined antibiotics, except colistin and ampicillin-sulbactam. Most of the isolates were resistant to antibiotics cefepime, ceftazidime, ceftriaxone, aztreonam, amikacin, gentamicin, and ciprofloxacin, especially to the three antibiotics cefepime, ceftazidime, and ceftriaxone. This finding indicates that colistin and ampicillin-sulbactam may be the most effective antibacterial agents against A. baumannii infections and of course better choices for their treatment. Similarly, in a recently published study by Mohajeri et al. on 104 samples of attending patients of Kermanshah hospital, they found that all isolates had high-level of resistance to imipenem and meropenem, while no resistance to polymyxin B, colistin, tigecylcine, and minocycline was observed (28). In a study by Rahbar et al., the resistance pattern of 88 strains of A. baumannii was studied from clinical specimens, and high rate of resistance to ceftriaxone (90.9%), piperacillin (90.9%), ceftazidime (84.1%), amikacin (85.2%), and ciprofloxacin (90.9%) was indicated (29).
In 2011, Vahdani et al. examined the resistance pattern of 101 strains of A. baumannii, isolated from hospitalized patients, and found that isolates were resistant to ceftazidime (96%), ceftizoxime (95%), ceftriaxone (93%), amikacin (58%), gentamicin (68%), co-terimoxazole (85%), and ciprofloxacin (85%) (30). Previously published experiments showed that A. baumannii strains are highly resistant to third and fourth generation of cephalosporins, carbapenems, quinolones, and aminoglycosides, such as cefepime, ceftazidime, ceftriaxone, aztreonam, amikacin, gentamicin, and imipenem, although some differences exist (31, 32). Variable resistance patterns in different populations might be partly due to hospital-associated healthcare settings available to patients, physician’s experience, medical care, and supportive care (33, 34).
As an example, in some studies, high percentage of resistance was related to cefotaxime, ceftazidime, ticarcillin, and piperacillin-tazobactam (31, 32). In this study, 30 phenotypic patterns were observed. Although each of these 30 isolates were resistant to at least seven antibiotics, one of these strains was resistant to 17 antibiotics, which is a remarkable observation in this study. It is necessary to mention that some of these patterns were comparable with other previously reported patterns in different countries or even Iran. For example, resistance to tobramycin and gentamicin in the current study was similar to the rates reported by previous studies in Iran (35, 36). The emergence of such MDR and PDR pathogens is a serious crisis in the medical community, an issue that could be solved through development of novel medications.
In the next step, ompA gene from these resistant strains was amplified using PCR. The ompA was considered as antigenic factor for production of the recombinant protein, due to its biological characteristics. The OmpA can attach to the eukaryotic cells, localized in the nucleus and promote apoptosis (7, 37). The OmpA can interact with complement factor H and inhibit immune responses (1). The amplified ompA was cloned and sub-cloned in pTZ57RT and in pET-32a(+) expression vector, respectively, and transferred to the E. coli host to produce rOmpA. The expressed recombinant OmpA was validated using SDS-PAGE analysis. This study is valuable as the recombinant OmpA protein was extracted from the resistant A. baumannii, found in the collected samples and not from commercially available standard strains. In a recent publication by Moriel et al., outer membrane proteins ompA and smpA, were reported as two of 62 potential candidates to develop recombinant vaccine against A. baumannii (19).
Additionally, OmpA, showed more than 99% similarity between all A. baumannii species, thus, it is a good candidate for vaccine development (38). In 2018, Ansari et al. produced a recombinant construct containing the ompA gene, which is notably expressed in HDF eukaryotic cells, and introduced this construct as a promising DNA vaccine candidate (2). Previously, Alzubaidi and Alkozai evaluated the potential of recombinant protein OmpA to study the cellular and humoral responses and found that rOmpA protects the rates against an infectious A. baumannii (11). Recently, a recombinant construct was developed to express the recombinant SmpA protein in prokaryotic cells. The SmpA is one the most important proteins for the localization of outer membrane proteins in the membrane (39). Although limited studies considered ompA as a recombinant vaccine, in the same approach, inactivated whole cell and outer membrane vesicles (OMVs) have been considered as target of vaccine generation against A. baumanii (40). As an example, recombinant proteins of Ata, Bap, and the K1 polysaccharide capsular have been produced and recognized as potential immunogens to conflict against A. baumannii infections (41-43).
Although the current researchers efficiently expressed the rOmpA in the prokaryotic system, there is a long way to achieve the recombinant protein vaccine. This includes extraction and purification of produced recombinant protein and studying its antigenic potential following injection in the mice experimental model. This is an issue, which should be addressed in the next experiments.
5.1. Conclusions
In this study, the antibiotic resistant feature of A. baumannii was revealed in 68 urinary tract infections, wound pus, septicemia, and septum. Among 30 isolates, 29 isolates were defined as MDR and one isolate was PDR. This findings showed the highly resistant pattern of A. baumannii in clinical samples. Consequently, the ompA gene from resistant isolates was extracted and cloned in a prokaryotic expression vector pET32a(+) to produce the recombinant OmpA protein. It has been observed that the recombinant OmpA protein was highly expressed in E. coli at both levels of transcript and encoded protein. The rOmpA may be a promising candidate for vaccine development, although further experiments are urgent. These include purification of recombinant rOmpA protein as well as immunological response assays in mice experimental model. The mentioned issues should be addressed in the near future. To the best of the author’s knowledge, this is the first report, in which the ompA gene was extracted from resistant A. baumannii isolates and cloned in expression plasmid to develop a recombinant protein, which can probably be used as vaccine in the near future, although further experiments are needed.