1. Background
Helicobacter pylori, a Gram-negative bacteria, is the most important cause of gastric ulcer, gastric malignancies (e.g., MALT lymphoma and gastric adenocarcinoma), and chronic gastritis. This bacteria infects half of the world's population (1-3), with an estimated prevalence of 70% - 90% and 35% - 40% in the developing and developed countries, respectively (4). Clinical symptoms are absent in the majority of infected patients, 10% - 20% of whom develop duodenal or gastric ulcers, and 1% present with gastric carcinoma (1, 5). The World Health Organization (WHO) in 1994 classified H. pylori as a type I carcinogen because of its role in gastric cancer (6-8). The effective elimination of the bacteria can be helpful for patients with gastric cancer and peptic ulcer. The gastric mucosal atrophy and inflammation reduce following the successful eradication of the bacteria, and the prevalence of H. pylori-associated peptic ulcer and gastric cancer reduces (9).
The eradication of H. pylori is normally achieved by combining a proton pump inhibitor with different antibiotics, including tetracycline, metronidazole, amoxicillin, and clarithromycin (10-12). Different resistance rates have been reported from different parts of the world. Therefore, the regional susceptibility pattern should be determined before any drug prescription for H. pylori infection (10, 13, 14). Although some factors can result in reduced efficacy of H. pylori treatments, antibiotic resistance seems to be the most important one (15). The resistance of H. pylori to many antibiotics rapidly increases following the treatment, thereby leading to partial bacterial elimination and rapid disease recurrence (11, 13).
Given its bactericidal effects on H. pylori, clarithromycin is recognized as the most important antibiotic for the treatment (12, 16, 17). Clarithromycin resistance is associated with the absence of drug binding to 23srRNA of the bacterial ribosome, related to point mutations in the variable domains of 23srRNA peptidyl transferase (PT). Although resistance is associated with different point mutations, the adenine-guanine transition at 2143s and 2142 (A2143G and A2142G) is the most important one (12, 16-19). While these mutations mostly account for clarithromycin resistance, each is independently associated with a specific minimum inhibitory concentration (MIC). The A2143G mutation has greater effects on the treatment outcomes compared with other mutations and is associated with a lower eradication rate (20, 21).
Metronidazole is an important drug for H. pylori treatment. It serves as a complement drug in quadruple, concomitant, hybrid, and sequential methods. The reduction of the nitro group in metronidazole increases the level of toxic free radicals resulting in DNA cleavage, helix instability, and cell death (22). The mechanism of resistance of H. pylori to metronidazole is quite complex and has a strong connection to the mutational inactivation of redox-associated rdxA and frxA genes (17, 18, 23); the most common cause of resistance is rdxA gene mutational inactivation. These mutations include insertion, deletion, frameshift, and missense mutations (18, 24). One of the most common mechanisms of resistance to metronidazole is 200-bp deletion mutation of the rdxA gene (25, 26).
Standard methods, including E-test, disk diffusion, agar dilution, and micro broth dilution, are used to evaluate H. pylori resistance in clinical specimens. Overall, for the measurement of MIC of most antibiotics, the E-test is considered an expensive but simple method. In comparison with the phenotypic methods, genotypic strategies, including sequencing, real-time PCR, and PCR-RFLP, are less time-consuming and can identify resistance-inducing mutations. However, considering the inadequate information about the mechanisms of resistance to some antibiotics, genotypic methods show limited efficiency (12, 17, 18). The PCR-RFLP assay, exhibits great efficiency in identifying 23s rRNA mutations (17).
2. Objectives
This study aimed to evaluate clarithromycin and metronidazole resistance in H. pylori and to detect point mutations in clarithromycin resistance as well as 200-bp deletion mutations in metronidazole resistance.
3. Methods
3.1. Sample Collection, Strains Isolation, Identification and Storage
All patients admitted to the endoscopy units of Mehr and Imam Hospitals of Ahvaz city during 23 July 2015 to 20 May 2016 entered the study. In total, 338 gastric biopsy samples were collected from patients. The samples were kept in sterile tubes containing BHI medium (Merck, Germany) and 5% non-activated fetal bovine serum (FBS; Baharafshan, Iran); then, they were transferred on ice to the microbiology department. After fragmenting and homogenizing the samples with a bistoury blade on a sterile glass slide, they were cultivated on Colombia agar (Merck, Germany), consisting of 5% inactivated FBS (Baharafshan, Iran), 7% fresh sheep blood, and antibiotics including trimethoprim (5 mg/L), amphotericin B (4 mg/L), polymyxin B (10 mg/L), and vancomycin (10 mg/L), all supplied by Sigma (USA). Next, the plates were incubated at a temperature of 37°C under microaerophilic conditions (saturated humidity; 5% oxygen and 7% Co2) for 3 - 10 days (Anaerocult C system; Merck, Germany) (27).
The urease, oxidase, and catalase tests, as well as Gram staining, were applied to identify the strains as H. pylori. Following that, BHI medium (Merck, Germany), consisting of 10% inactivated FBS and 30% sterile glycerol, was used to prepare a bacterial suspension; then, it was stored at -80°C (27). The patients completed a questionnaire before endoscopy. Patients who used any antibiotics for 14 days prior to the endoscopy were excluded from study.
3.2. DNA Extraction
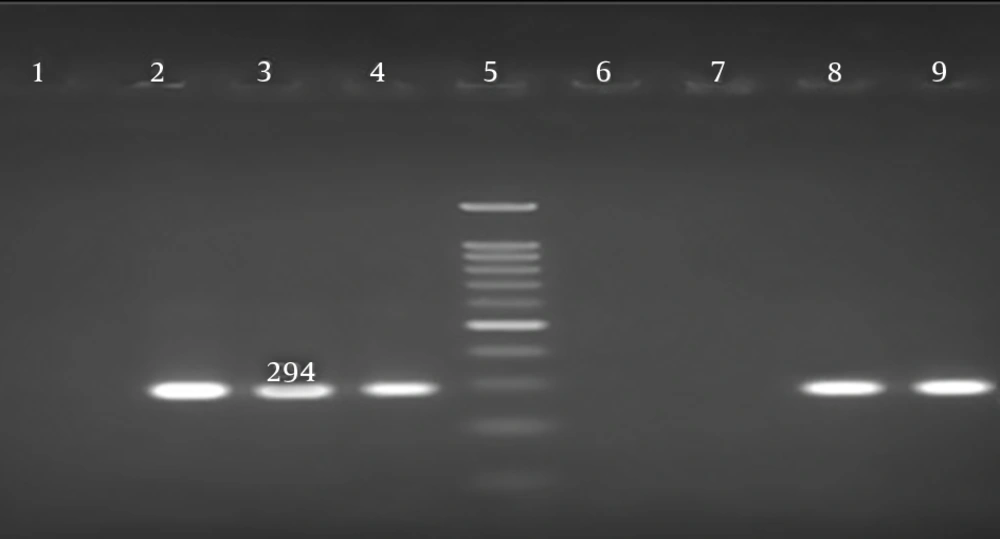
A DNA extraction kit (Roche, Germany) was used to extract the DNA from the H. pylori isolates, as outlined by the manufacturer and the extracted DNA was stored at a temperature of -20°C (8, 12). To confirm H. pylori isolation, PCR assay was performed to amplify glmM (ureC) gene, producing a 294-bp fragment; this housekeeping gene can be identified in all H. pylori strains. The PCR assay included denaturation for 5 minutes at 94°C, followed by 40 cycles of denaturation for 1 minute at 94°C, annealing for 90 seconds at 57°C, and extension for 2 minutes at 72°C. An extension was applied at 72°C for 7 minutes to complete the PCR assay. The Master Mix (total volume, 20 µL; Sinaclon, Iran) consisted of 800 µmol of dNTPs, 2.5 µg of MgCl2, 50 µm of buffer, 0.7 µL of template DNA, 2.5 µg of Taq enzyme, and 1.8 µL of each primer (Metabion, Germany). Ethidium bromide was used to electrophorese the products on 1.5% agarose gel (28, 29). Table 1 shows the primer sequences and names, genes, and size of the PCR products.
3.3. Detection of Clarithromycin and Metronidazole Resistant H. pylori Using E-Test
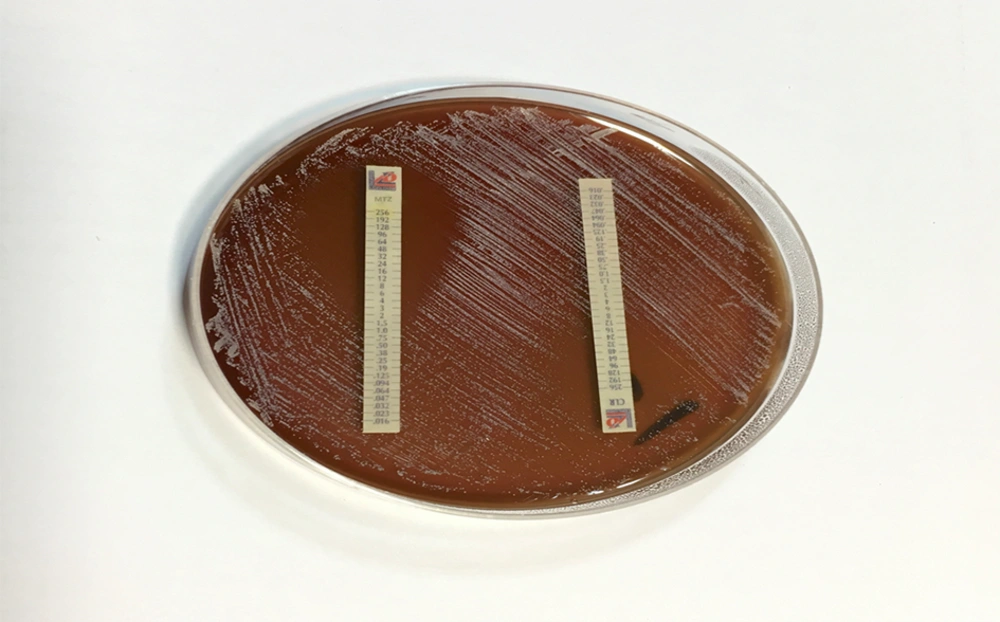
The E-test was applied in the antibiogram test for H. pylori strains, using strips of clarithromycin and metronidazole (Lioflichem, Italy) on Mueller-Hinton agar (MHA; Merck, Germany) which was enriched with 7% sheep blood. In all the strains, the MICs of metronidazole and clarithromycin were measured. A suspension containing pure H. pylori colonies was first prepared using sterile physiological saline (9 × 108 CFU/mL; turbidity, 3 McFarland) and inoculated on MHA with a suspension-immersed cotton swab. The strips were placed on the medium after drying the medium surface; they were then incubated under microaerophilic conditions for 72 hours at 37°C. MICs ≥ 1 and 8 µg/mL were considered resistant to clarithromycin and metronidazole, respectively (27).
3.4. Amplification of the Variable Domains of PT from 23s rRNA Gene
In the PCR, to amplify the variable domain of 23srRNA PT, initial denaturation was performed at 94°C for 4 minutes, followed by 32 cycles of denaturation at 94°C for 40 seconds, annealing for 1 minute at 61.5°C, and extension for 1 minute at 72°C; an extension at 72°C for 7 minutes ended the reaction. The Master Mix (total volume, 25 µL; Sinaclon, Iran) included 2.5 µL of dNTPs, 3.3 µL of buffer, 0.2 µL of Taq enzyme, 0.7 µL of template DNA, and 1.8 µL of each primer (Metabion, Germany). Ethidium bromide (Sinaclon, Iran) was used to electrophorese the PCR product on 1.5% agarose gel (30). Table 1 presents the sequence and name of primers, name of genes, and size of PCR products.
3.5. Detection of Point Mutations via PCR-RFLP Assay in Strains Resistant to Clarithromycin
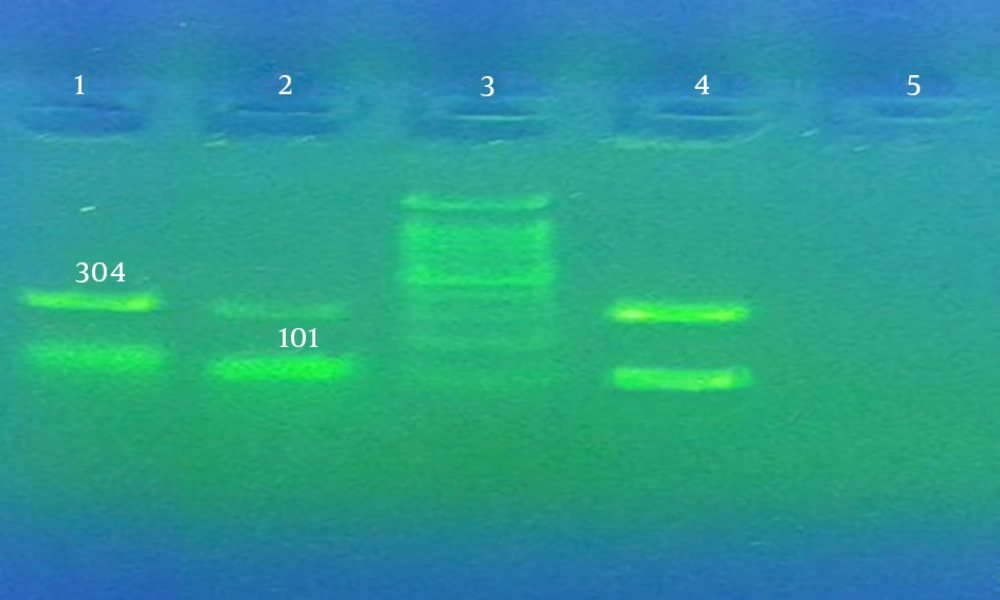
The PCR-RFLP assay was performed in clarithromycin-resistant strains to identify point mutations. PCR was first applied to amplify a fragment (425 bp) from the 23s rRNA PT variable domain. Then, BsaI and BbsI restriction enzymes (Thermoscientific, Lithuania) were used to digest the PCR products. Electrophoresis was performed on 1.5% agarose gel to detect the fragments. In the RFLP method, point mutations in clarithromycin resistance were identified in the resistant strains based on the size and number of fragments (11, 16, 23, 24). The mutations were detected via sequencing using an ABI system (Prism 310 Analyzer, USA), in case other point mutations were present. For enzymatic digestion of the PCR product from amplification of PT variable region, 8 µL of distilled water was mixed with the PCR product (2 µL) and digestive enzymes (2 µL). Incubation was performed at 37°C for 30 minutes and 37°C for 24 hours in BsaI and BbsI enzymes, respectively. Then, electrophoresis was conducted on the digested products (11, 15, 16).
3.6. Identification of 200-bp Deletion Mutation Responsible for Metronidazole Resistance
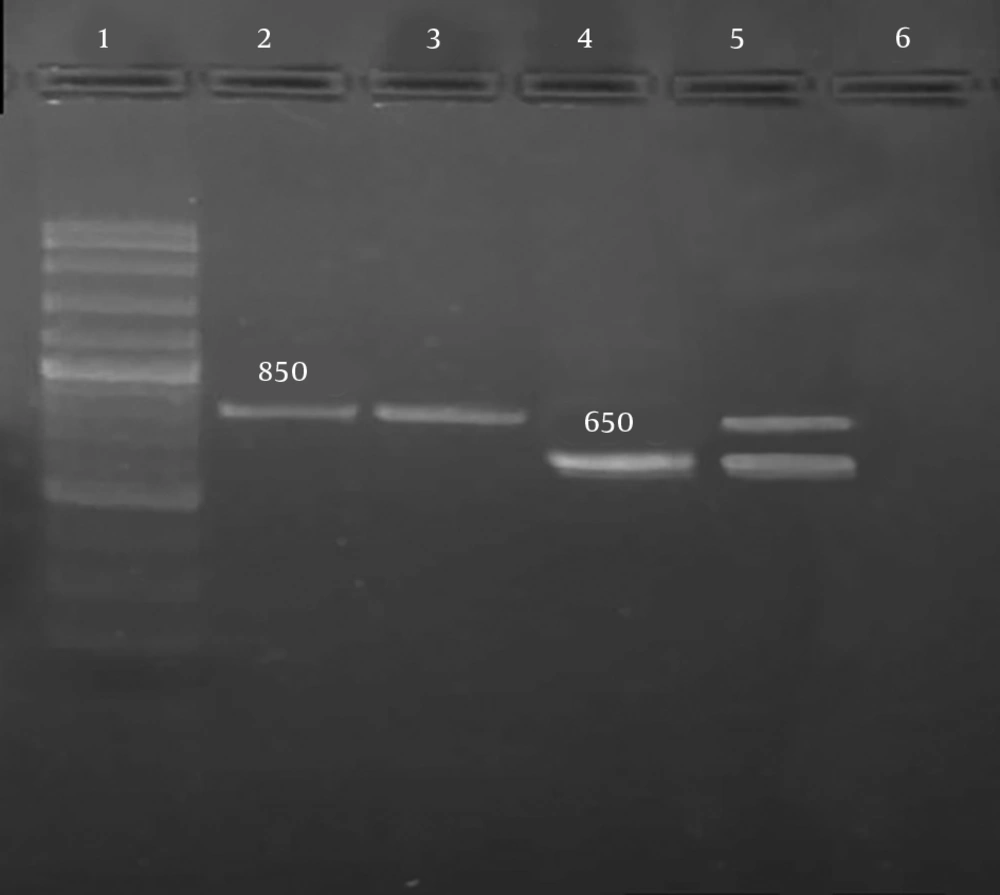
To determine one of the molecular mechanisms of resistance to metronidazole, the PCR method and primers specified in Table 1 were used. Deletion of a 200-bp fragment from the rdxA gene was investigated. The removal of this fragment leads to resistance to metronidazole. If the mutation does not occur and rdxA gene is intact, amplification of this gene by PCR leads to the production of an 850-bp fragment; if the mutation occurs, amplification of the defective gene by PCR leads to the production of a 650-bp fragment.
The PCR assay included denaturation for 5 minutes at 95°C followed by 30 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extension at 72°C for 1 minute; a final extension (5 minutes, 72°C) ended the reaction. The Master Mix (total volume, 25 µL; Sinaclon, Iran) consisted of 800 µmol of dNTPs, 50 mmoL of buffer, 2.5 units of Taq enzyme, 2.5 mmol of MgCl2, 1 µL of template DNA, and 0.25 µL of each primer (Metabion, Germany). Then, using ethidium bromide (Sinaclon, Iran), the products were electrophoresed on 1.5% agarose gel (23). Table 1 presents the sequence and name of primers, size of PCR products, and gene names.
3.7. Statistical Analysis
Because the aim of this study was to identify the mutations responsible for resistance to clarithromycin and metronidazole, No statistical methods were used in this article.
4. Results
In total, 131 (38.7%) H. pylori strains were cultured from the 338 endoscopic samples. In total, 70 (53.4%) and 61 (46.6%) strains were isolated from men and women, respectively. As the findings revealed, men and women were within the age range of 16 - 84 and 24 - 90 years, respectively. The grown colonies of the H. pylori were very fine, gray, transparent and relatively convex. Spiral Gram-negative strains, positive for urease, oxidase and catalase tests, were initially identified as H. pylori. PCR assay was performed for the amplification of UreC gene (glmM) and confirming the H. pylori strains (Figure 1). The clinical diagnoses based on endoscopy (confirmed via pathology) included gastric cancer, duodenal ulcer, peptic ulcer, and gastritis.
Among the 131 H. pylori strains, 70 (53.4%) and 83 (63.4%) showed resistance to clarithromycin and metronidazole, respectively, based on E-test (Figure 2). The MIC was 1 - 96 µg/mL in clarithromycin-resistant strains whereas the susceptible strains had MICs below 1 µg/mL. On the other hand, the MIC was 8 - 128 µg/mL in metronidazole-resistant strains, and the susceptible H. pylori strains showed MICs below 8 µg/mL.
To detect point mutations in clarithromycin resistance, PCR was performed for amplifying a fragment (425 bp) from 23srRNA PT variable domain. The BsaI and BbsI endonuclease enzymes affected the PCR product. The PCR product of the strains containing A2143G mutation produced 304-bp and 101-bp fragments if affected by BsaI (Figure 3) while the strains containing A2142G mutation produced 332-bp and 93-bp fragments if affected by BbsI. As the findings revealed, 425-bp fragments were produced by the clarithromycin-resistant strains on the PCR assay.
For the detection of deletion mutation responsible for the resistance to metronidazole, PCR was conducted, and an 850-bp fragment of the rdxA gene was amplified. Metronidazole-resistant strains containing 200-bp deletion in the rdxA gene produced 650-bp fragments while the strains containing intact rdxA genes produced 850-bp fragments (Figure 4).
Among the clarithromycin-resistant isolates (n, 70) affected by restriction enzymes, 50 (71.4%) and 5 (7.1%) strains included A2143G and A2142G mutations, respectively; also, 11 (15.7%) strains contained both A2142G and A2143G mutations. Sequencing was carried out in 4 strains which lacked mutations and were clarithromycin-resistant on the E-test. The sequences were examined using MEGA7 version 7 (2016, for bigger datasets). A2142C mutations were present in 3 strains; one strain had no mutations. Seven (8.4%) out of the 83 strains, which were resistant to metronidazole, included 200-bp deletion whereas all showed resistance based on the E-test. Also, a standard strain of H. pylori ATCC26695 was used as the control.
5. Discussion
Helicobacter pylori resistance to antibiotics is expanding worldwide, and Iran is among the countries where the antibiotic resistance is spreading. Clarithromycin is extensively used in H. pylori treatment; therefore, increased resistance has become a major issue in the eradication of the bacteria (31). It is difficult to apply culture-based phenotypic methods to evaluate H. pylori sensitivity to drugs, as these bacteria are fastidious and require specific atmospheric conditions for growth on selective media for long periods (32, 33). Helicobacter pylori resistance to clarithromycin and metronidazole varies worldwide. Resistance to clarithromycin has been reported to be 30% in Southern Europe and 15% - 30% in the United States (19). In Asian countries, such as Pakistan and Iran, resistance has been estimated at 47.8% and 31.7%, respectively (31, 34). Resistance to metronidazole also varies in different parts of the world. The resistance rate has been reported to be very high in some regions of Asia and Africa and low in some others. For example, the resistance rate has been reported to be 77.8% and 68.8% in China and Gambia, respectively (12, 35). Low rates of metronidazole resistance, 23.9% and 40.5%, have been reported from Taiwan and Iran, respectively (36, 37).
Based on the E-test method, the isolated H. pylori strains showed resistance to clarithromycin in 70 out of 131 (53.4%) cases in this study, which is in line with the studies by De Francesco et al. (13) from Italy (51.2%) and Rasheed et al. (34) from Pakistan (47.8%). However, the results are largely incompatible with the studies by Lottspeich et al. (38) from Germany (13.3%) and Tanih and Ndip (10) from South Africa (15.4%). Moreover, Caliskan et al. from Turkey reported the resistance to clarithromycin to be 36.7%, which is somewhat close to the resistance rate in this study (39). Different resistance rates of H. pylori to clarithromycin have been reported in different regions of Iran. In studies by Safavi et al. (40) and Shokrzadeh et al. (37) the resistance rate was reported to be 22.4% and 14%, respectively (37, 40), whereas Abdollahi et al. reported a rate of 31.7% (31). For the first time, Versalovich and colleagues reported that point mutations in the 23s rRNA variable region are related to the resistance of H. pylori to clarithromycin. Point mutations, which result in adenine-to-guanine transition in 23s rRNA sequence, can be found at positions 2143 and 2142 (31).
In this study, A2143G and A2142G mutations were found in H. pylori clarithromycin-resistant strains, based on the PCR-RFLP assay; other mutations were detected via sequencing when these mutations were not present. At least one point mutation was present in all the resistant strains, and the correlation of clarithromycin resistance in H. pylori with point mutations was confirmed. The A2143G and A2142G mutations were detected in 71.4% and 7.1% of the strains, respectively, in this study. This finding is in line with the results reported by Agudo et al. (16) from Spain, Doroud et al. (23) from Iran, and Nishizawa and Suzuki (41) from Japan. However, the findings are inconsistent with the results of the studies by Wolle et al. (11) from Germany and Abdollahi et al. (31) from Iran. Therefore, there are geographical differences in clarithromycin resistance which highlights the significance of identifying regional patterns of resistance in H. pylori for the selection of proper treatment. The mutations discussed in this study are not the only established H. pylori point mutations in clarithromycin resistance, and there are reports of some other mutations worldwide. Moreover, some of the clarithromycin resistance is attributed to the efflux system in H. pylori, which forces the bacteria to extrude the macrolides.
Gudwin et al. were the first researchers to show that mutational inactivation of rdxA gene leads to metronidazole resistance. One of these mutations is the 200-bp deletion of rdxA gene. In the present study, this type of mutation was investigated. There is a debate regarding the deletion mutation among researchers. While Ossenkopp et al. showed that this type of mutation is a major factor in metronidazole resistance, Kato et al. reported that they did not find any 200-bp deletion in metronidazole-resistant strains. Various contradictory results have been obtained in many studies from Iran. While Mohammadi et al. and Kargar et al. emphasized on the presence of 200-bp deletion among the resistant strains, Mirzaei et al. (42) did not report this type of mutation. Doroud et al. (23) reported the presence of 200-bp mutation in 5% of resistant strains, which is close to the reported rate in this study. However, Abdollahi et al. (31) reported the prevalence of this type of mutation in 22.9% of resistant strains, which is different from the results of the present study. Regarding the 200-bp deletion mutation, the present results revealed that this mutation accounted for a small part of H. pylori resistance. Other mechanisms of H. pylori resistance appear to be involved in metronidazole resistance of the studied strains.
5.1. Conclusions
The high prevalence of resistance to clarithromycin and metronidazole in H. pylori is a major concern revealed by this study which should be taken into account by physicians in selecting drug regimens. The results confirmed the necessity of phenotypic and genotypic methods of antibiotic susceptibility. The association between H. pylori resistance to clarithromycin and 23srRNA point mutations was confirmed. Based on the findings, H. pylori resistance to clarithromycin and metronidazole has been increasing in Iran, resulting in the reduced effectiveness of these antibiotics. The high H. pylori resistance to metronidazole and clarithromycin may be associated with the prevalent use of clarithromycin for respiratory infections and metronidazole for parasitic, gynecologic, and oral infections in Iran. Also, another reason for the increased resistance rate could be the prevalent use of clarithromycin and metronidazole in H. pylori treatments due to the high frequency of gastric disorders in the study population.