1. Background
Hepatitis C virus (HCV) is a globally prevalent pathogen and the top trigger of death and morbidity (1). Hepatitis C virus prevalence in the last 15 decades have been shown to raise to 2.8%, estimated to be equal to > 185 millions of infection worldwide (2). Chronic HCV infection may possibly progress to severe liver disease, which includes cirrhosis and at some point, into hepatocellular carcinoma (3). Annually 3% - 4% of population is infected and thousands of deaths occur worldwide due to hepatitis C. Among infected individuals, 85% of sufferers advance into chronic stage and then to cirrhosis, 1% - 4% of the patients lead to hepatocellular carcinoma (HCC) (4).
On the basis of phylogenetic and sequence evaluation, HCV has been classified into one to seven key genotypes (5). These strains differ from one another at 30% - 35% nucleotide sites. These genotypes have been further classified into 67 confirmed and 20 provisional subtypes and these are distinct at < 15% of the nucleotide sites (6). Geographical distribution of HCV genotypes is extremely complex, it has been known that genotypes known as epidemic genotypes, 1, 2, and 3 have worldwide distribution and account for a massive proportion of HCV infection in well developed countries (7-9). Other genotypes called as endemics are comparatively rare and are present in quite restricted regions. Endemic strains from genotype 1 and 2 are present in West Africa, 3 in South Asia, 4 in Central Africa and Middle East, 5 in Southern Africa, and 6 in South East Asia (10). Genotype 7 has been isolated in Canada from Central African immigrants (11).
Distinct techniques have been utilized to figure out genotypes, amongst the most frequently employed are, direct sequencing of a distinct polymerase chain response (PCR)-amplified portion of the viral genome obtained from a patient sample, followed by phylogenetic examination restriction fragment length polymorphism (RFLP) (5, 12). PCR amplification, with variety-distinct primers, has been used to figure out HCV genotype, however, sensitivity and specificity parameters of this approach demand even more evaluation (4, 12, 13). DNA hybridization technologies, including LiPA, are commercially offered to figure out HCV genotypes (13). Lastly, kinetic PCR with fluorescence resonance vitality transfer (FRET) probes or intercalating dyes has been utilized to detect genotype HCV in a single tube reaction utilizing dissociation curve analysis (14, 15).
Everywhere in the world treatment strategies are based on HCV genotypes, therefore, prior to therapy, it’s essential to determine particular genotype for the sake of deciding type and duration of antiviral therapy either direct-acting antiviral agents (DAA) or indirect acting antiviral agents (AA). In Pakistan, HCV prevalence is increasing day by day and it is almost in the range of 3% - 5% (16), depending on the regions within Pakistan. Here most of the people determine HCV genotype using Ohno et al. (17), HCV genotyping method. However, discrepancy exists in genotyping. As some time different laboratories issue different genotypes or sub types of the same sample. This might be due to the in-house methods that have been developed. Secondly, treatment response has been linked with major HCV genotypes.
2. Objectives
In order to minimize these discrepancies, we employed real time PCR based genotyping method, based on principle of Florence hybridization (TaqMan Probe assay).
3. Methods
3.1. Ethics Statement
This study was approved by the Ethical Committee of Department of Biotechnology of Abdul Wali Khan University Mardan and Mercy Teaching Hospital, Peshawar, Khyber Pakhtunkhwa, reference number Awkum- 125.
3.2. Patients’ Selection and Blood Collection
This study included 100 patients. Patients were selected who were referred by physicians, either with relapse history or naïve patients. From each patient, 5 mL of blood was taken with informed consent. Blood was centrifuged at 8000 rpm for 5 minutes and serum was stored at -4°C until further use.
3.3. RNA Extraction
RNA extraction was done as previously performed (16). Briefly, the following procedure, according to manufacturer’s instructions (Macherey-Nagel GmbH, Germany), was followed. A total of 200 uL lysis solutions was taken in an eppendorp tube, added 200 uL serum sample, mixed it with the help of shaking, added 280 uL ethanol into the mixture, and transferred all of the above mixture into the receiver tube having spin column. After centrifugation, the receiver tube was discarded and transferred the spin column to another receiver tube, washed twice with the washing solution of the spin column, and finally 60 uL of elution buffer was added into the spin column and after slight centrifugation, HCV RNA was obtained.
3.4. Genotype Determination
To find out the genotype in each of the subject sample, Sacace real time PCR based genotyping kit (Sacace, Italy) was used. For each of the subjects, genotyping was done according to manufacturer’s instruction. Simply, master mix was prepared as shown in Table 1. All the above ingredients were mixed and added to specialized PCR tube, 15 uL master mix along with 10 uL extracted RNA (total volume of the mix was equal to 25 uL). Similarly, 10 uL controls, HCV genotype 1, 2, and 3 and dH20 was added to the tubes labeled as positive and negative control, respectively, and centrifuged slightly; all PCR tubes were placed in the PCR machine, Smart Cycler (Cepheid, USA). Set thermal protocols (Table 2) designated the sites and started the run. After two hours, the results and analysis were given and their interpretation is given in Table 3. The above procedure was adopted for all of the 100 samples. Positive and negative controls were added for validation of the results.
| Reagents | Volume in uL/Sample |
|---|---|
| RT-Mix 1 FRT | 10 |
| RT-Mix 11 | 0.5 |
| Taq polymerase | 0.5 |
| Total | 15.5 |
| Time in Cycle, Min | Temperature, °C | Detection |
|---|---|---|
| 1 (Hold) | ||
| 30 | 50 | |
| 15 | 95 | |
| 5 Repeats | ||
| 5 seconds | 95 | |
| 20 | 60 | |
| 15 | 72 | |
| 40 Repeats | ||
| 10 | 95 | |
| 30 | 60 | Floresence detection FAM. CY3, Texas Red and CY5 |
| 20 | 72 |
| Type of Channels | Interpretation |
|---|---|
| FAM | Genotype 1 |
| Cy3 | Genotype 2 |
| Texas Red | Genotype 3 |
| Cy5 | Amplification |
4. Results and Discussion
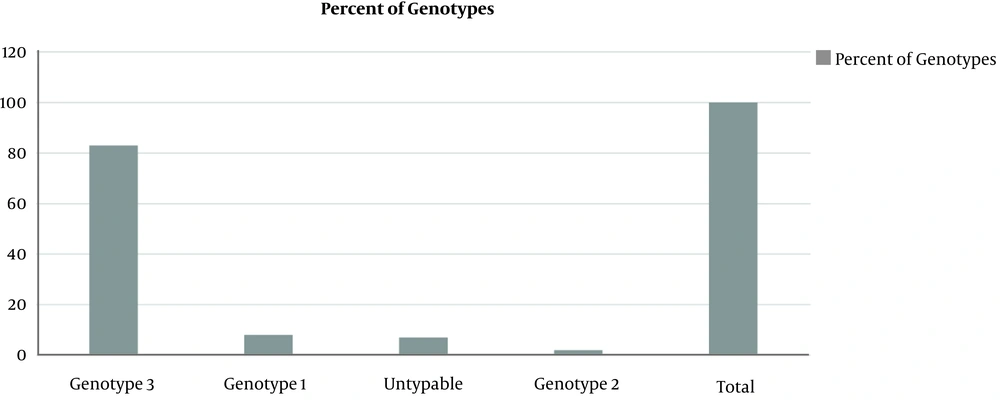
All sera samples were processed for genotyping using commercial Sacace real time PCR kit. The primer and probe was designed using 5 UTR region of HCV genome. The probe was tagged with FAM, CY3, and CY5. Interpretation of results is given in Table 3. While the percent prevalence is out of 100 subject population, the major genotype found was 3 (83%), followed by genotype 1 (8%) and untypable (7%) while the least genotype was 2 (2%) (Figure 1). While no amplification was found in negative control.
Chronic viral hepatitis and its associated complications with progressive liver disease are severe health problem worldwide. In Pakistan, its prevalence varies in different geographical regions and may range from 2% - 7% (18). Thousands of mortality is the result of chronic HCV infection (7). This wide spread is mostly associated with no/ low awareness regarding the normal procedures adopted for the diagnosis and treatment (19). HCV has been classified based on sequence analysis into distinct clades, genotypes, and subtypes (20). These genotypes have distinct worldwide distribution such as genotype 1 and 3 has worldwide distribution (10).
This study was conducted to figure out the percent prevalence of major HCV types using the Real-Time-based typing system (Sacace, Italy) for the first time in the Khyber Pakhtunkhwa province of Pakistan. The results indicated that major HCV genotype was type 3 (83%), followed by type 1 (8%) while a number of cases were untypeable as well (Figure 1). Previous studies in Pakistan, and even in Khyber Pakhtunkhwa, have already reported higher prevalence of the genotype 3a (16), which seem in line with our study. However, prevalence of genotype 1 is comparatively higher than the previous studies. This higher percentage indicates a new shift in HCV epidemiology, although the presence of genotype 1a and 1b has been reported by other studies conducted in Pakistan (16). A more recent study conducted in the Khyber Pakhtunkhwa province has also reported a shift the distribution of HCV types with higher percentage of HCV 1b or its mixed infections (21).
Presence of the ‘untypeable’ genotypes (Figure 1) has also been shown by other studies (16) however, its share in our study (7%) is higher than the previous reports. This might be due to low viral titer in the blood of the patients and absence of viral RNA in the serum samples or limitations of the present assay used, as this assay could detect only major genotypes 1 - 3. Prevalence of genotype 2 (Figure 1) has also been shown in studies (22), however, the percentage of existence of this particular genotype shown in this study is lower than previous studies (1, 16, 23).
4.1. Conclusions
Although a significant prevalence of major genotype 3 was found in the studied population, the overall pattern of HCV major types distribution indicates a shift in their distribution in contrast to majority of the studies conducted earlier.