1. Background
Due to the weakness and suppression of the immune system of hospital patients, hospital food should have acceptable hygienic safety. Amongst all known pathogenic bacteria responsible for food-borne disorders especially in public places, such as hospitals, Escherichia coli had an imperative role (1, 2). It is mainly responsible for outbreak of food poisonings in public places, including hospitals (1, 2). Escherichia coli are Gram-negative, non-sporulating, motile, flagellated, and rod shape bacteria with ability of growth in different types of foods under aerobic and anaerobic conditions. Shiga (vero) toxin (Stx) Escherichia coli (STEC) is a division of a noteworthy virulent group of the mentioned bacterium, known as enterohemorrhagic Escherichia coli (EHEC) (1, 3-5). Shiga toxin E. coli bacteria are responsible for rigorous medical disorders, such as hemorrhagic colitis (HC), lethal hemolytic uremic syndrome (HUS), different kinds of diarrhea, and thrombotic thrombocytopenic purpura (TTP) (1, 4).
Outburst of food disorders are mainly related to certain serogroups and particularly O157 (1, 2, 4-6). This serogroup is responsible for severe cases of food-borne gastrointestinal disorders (3-6). Some imperative virulence genes comprising intimin (eaeA), Shiga toxins (stx1 and stx2), and hemolysin (hlyA) are responsible for occurrence of E. coli colonization, adhesion, and invasion to intestinal walls (3-6). High prevalence of resistance in E. coli O157 bacteria is an alternative imperative factor, which enhances the pathogenicity of bacteria. Inappropriately, O157 E. coli bacteria related to food and clinical implications exhibited a high prevalence of resistance against normally used antibiotics (50% to 100%), especially tetracyclines, aminoglycosides, fluoroquinolones, macrolides, phenicols, and β-lactams (2-8).
2. Objectives
Regarding the indeterminate role of E. coli O157 bacteria in foods of hospitals and lack of epidemiological surveys in this area in Iran, the current examination was performed to assess the frequency of virulence genes and phenotypic analysis of antibiotic resistance of E. coli O157 bacteria isolated from gavage and soup hospital food samples.
3. Methods
3.1. Samples
From April to August 2016, a total of 200 hospital food samples, such as gavage (n = 100) and soup (n = 100), were randomly collected from educational hospitals of Iran. Samples were directly transported to the laboratory in chillers with ice-packs. All gavage and soup samples displayed usual physical features and organoleptic characters.
3.2. Escherichia coli O157 Isolation and Identification
For each sample, 25 g was homogenized and 1 g of the homogenate was added to 5 mL of buffered peptone water (Merck, Germany) and incubated. Cultures were speckled onto MacConkey sorbitol agar (Merck, Germany) and plates were incubated instantly at 37°C. From each plate, five to ten distrusted colonies of E. coli bacteria (with both positive and negative sorbitol activities) were chosen and cultured another time on likely medium. Latex agglutination method was used for examination of O157 antigen in the sorbitol-negative bacteria (Oxoid, Uk) (9). Isolates were further identified using biochemical tests, including Gram staining, cytochrome oxidase, Indole test, Methyl Red test, Voges-Proskauer test, citrate utilization (IMViC) test, and lysine decarboxylase test.
3.3. Polymerase Chain Reaction Detection of Virulence Factors
Bacteria were cultured another time in Luria-Bertani broth (Merck, Germany) at 37°C. DNA was extracted from bacteria by DNA extraction kit (Thermo Fisher Scientific, Germany) rendering to the producer’s guidelines. Table 1 displays the oligonucleotide primers and PCR circumstances applied for amplification of virulence genes (10, 11). DNA thermo-cycler (Flexrcycler2, Germany) was applied for this purpose. Fifteen microliters of the PCR products were introduced to 1.5% agarose gel electrophoresis (4).
| Target Gene | Primer Sequence (5’- 3’) | PCR Product (bp) | PCR Programs | PCR Volume (50 µL) |
|---|---|---|---|---|
| stx1 | F: AAATCGCCATTCGTTGACTACTTCT | 366 | 1 cycle: 95°C (3 min); 34 cycle: 94°C (60 s), 56°C (45 s), 72°C (60 s); 1 cycle: 72°C (10 min) | 5 µL PCR buffer 10X; 2 mM Mgcl2; 150 µM dNTP (Fermentas); 0.75 µM of each primers F & R; 1.5 U Taq DNA polymerase (Fermentas); 3 µL DNA template |
| R: TGCCATTCTGGCAACTCGCGATGCA | ||||
| stx2 | F: CGATCGTCACTCACTGGTTTCATCA | 282 | ||
| R: GGATATTCTCCCCACTCTGACACC | ||||
| eaeA | F: TGCGGCACAACAGGCGGCGA | 629 | ||
| R: CGGTCGCCGCACCAGGATTC | ||||
| ehly | F: CAATGCAGATGCAGATACCG | 432 | ||
| R: CAGAGATGTCGTTGCAGCAG |
3.4. Antimicrobial Susceptibility Testing
Mueller-Hinton agar (Merck, Germany) was applied for assess the pattern of antibiotic resistance using the simple disk diffusion technique. Antibiotic resistance of E. coli O157 bacteria was studied against mezlocillin (30 u), imipenem (30 u), ampicillin (10 u), cefotaxime (30 µg), levofloxacin (5 µg), gentamycin (10 µg), ceftazidime (30 µg), cotrimoxazole (30 µg), cefepime (30 µg), enrofloxacin (5 µg), ciprofloxacin (5 µg), sulfamethoxazole (25 µg), tetracycline (30 u), trimethoprim (5 µg), and chloramphenicol (30 µg) (Oxoid, UK), using the Clinical and Laboratory Standards Institute guidelines (12).
3.5. Statistical Analysis
Data obtained from the experiments were transferred to Excel software. SPSS/21.0 software was applied for statistical analysis. Chi-square test and Fisher’s exact two-tailed test were used to assess any significant relationship between parameters of the study. Numerical meaning was regarded at a P value < 0.05.
4. Results
Table 2 characterizes the distribution of E. coli O157 in hospital food samples. Nine out of 200 (4.50%) hospital food samples were positive for E. coli O157. Distribution of E. coli O157 in soup and gavage samples was 3% and 6%, respectively. A statistically significant difference was observed for the distribution of E. coli O157 bacteria between soup and gavage samples (P < 0.05).
| Types of Samples | No. Samples Collected | No Positive Strains (%) |
|---|---|---|
| Soup | 100 | 3 (3) |
| Gavage | 100 | 6 (6) |
| Total | 200 | 9 (4.50) |
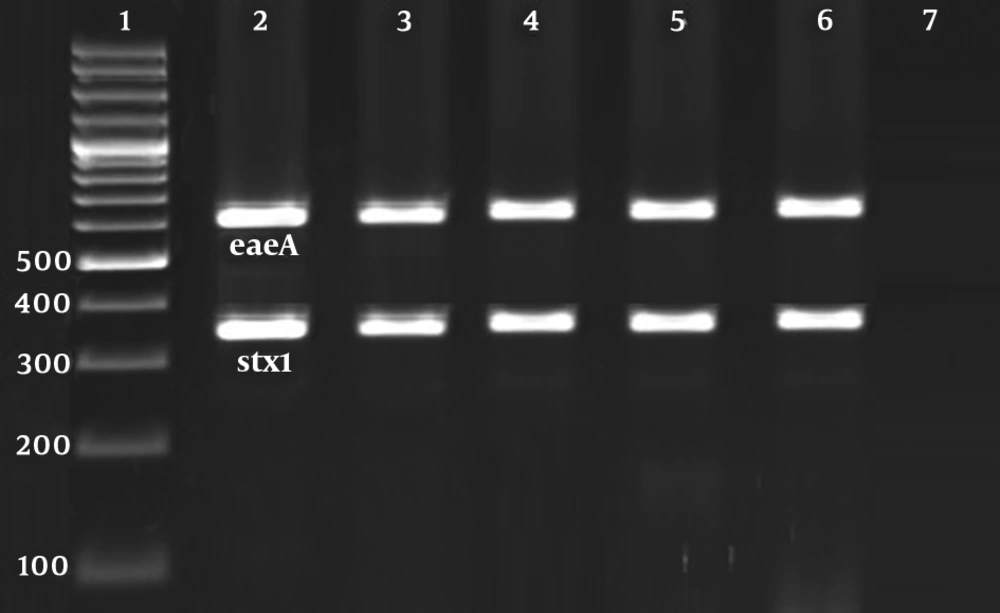
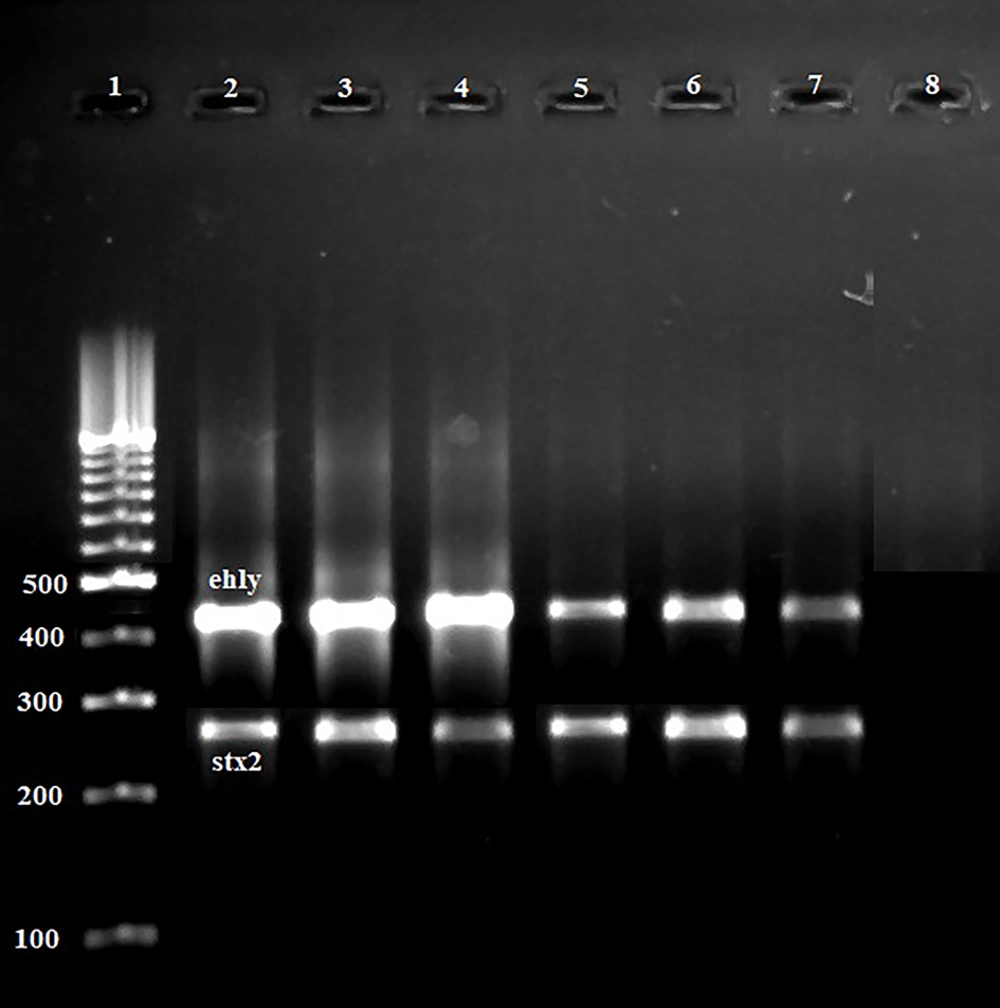
Figure 1 and 2 represent the results of the gel electrophoresis of PCR products for studied virulence factors. Table 3 characterizes the distribution of virulence genes amongst the E. coli O157 bacteria isolated from hospital food samples. Stx1 (100%), eaeA (100%), and ehlyA (100%) were the most routinely identified virulence genes amongst the E. coli O157 bacteria isolated from hospital food samples. Distribution of stx2 virulence gene was 22.22%. Escherichia coli O157 bacteria isolated from soup samples had a higher distribution of stx2 gene (P < 0.05). All isolates were simultaneously positive for stx1, eaeA, and ehly genes (100%).
Results of the gel electrophoresis of PCR products for virulence factors. 1: 100 bp ladder (Thermo Fisher Scientific, Germany). 2 - 6: Positive samples for eaeA (629 bp) and stx1 (366 bp) virulence factors. 7: Positive control for each gene and 8: Negative control (Water PCR buffer (Thermo Fisher Scientific, Germany)).
Results of the gel electrophoresis of PCR products for virulence factors. 1: 100 bp ladder (Thermo Fisher Scientific, Germany). 2 - 6: Positive samples for ehly (432 bp) and stx2 (282 bp) virulence factors. 7: Positive control for each gene and 8: Negative control (Water PCR buffer (Thermo Fisher Scientific, Germany)).
| Types of Samples | No. E. coli O157 Positive Samples | Distribution of Virulence Genes (%) | ||||
|---|---|---|---|---|---|---|
| stx1 | stx2 | eaeA | ehly | stx1+eaeA+ehly | ||
| Soup | 3 | 3 (100) | 1 (33.33) | 3 (100) | 3 (100) | 3 (100) |
| Gavage | 6 | 6 (100) | 1 (16.66) | 6 (100) | 6 (100) | 6 (100) |
| Total | 9 | 9 (100) | 2 (22.22) | 9 (100) | 9 (100) | 9 (100) |
Table 4 characterizes the antibiotic resistance pattern of E. coli O157 bacteria isolated from hospital food samples. Escherichia coli O157 bacteria exhibited maximum incidence of resistance against tetracycline (100%), gentamycin (100%), ampicillin (100%), mezlocillin (50%), enrofloxacin (50%), and trimethoprim (50%) antibiotics. Distribution of resistance against imipenem (11.11%) and chloramphenicol (22.22%) was lower than other tested antibiotic agents. A statistically significant difference was observed for the distribution of antibiotic resistance between soup and gavage samples (P < 0.05).
| Types of Samples (No. O157 Positive Samples) | Antibiotic Resistance Pattern (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Teta | Cfz | Mez | Cfx | Gen | Cot | Enr | Cip | Imp | Amp | Tri | Lev | C30 | |
| Soup (3) | 3 (100) | 1 (33.33) | 1 (33.33) | 1 (33.33) | 3 (100) | 2 (66.66) | 3 (100) | 1 (33.33) | - | 3 (100) | 2 (66.66) | - | 2 (66.66) |
| Gavage (6) | 6 (100) | 2 (33.33) | 4 (66.66) | 3 (50) | 6 (100) | 3 (50) | 1 (16.66) | 3 (50) | 1 (16.66) | 6 (100) | 3 (50) | 3 (50) | - |
| Total (9) | 9 (100) | 3 (33.33) | 5 (55.55) | 4 (44.44) | 9 (100) | 5 (55.55) | 4 (44.44) | 4 (44.44) | 1 (11.11) | 9 (100) | 5 (55.55) | 3 (33.33) | 2 (22.22) |
aTet: tetracycline (30 u/disk), Cfz: ceftazidime (30 µg/disk), Mez: mezlocillin (30 u/disk), Cfx: cefotaxime (30 µg/disk), Gen: gentamycin (10 µg/disk), Cot: cotrimoxazole (30 µg/disk), Enr: enrofloxacin (5 µg/disk), Cip: ciprofloxacin (5 µg/disk), Imp: imipenem (30 u/disk), Amp: ampicillin (10 u/disk), Tri: trimethoprim (5 µg/disk), Lev: levofloxacillin (5 µg/disk), C30: chloramphenicol (30 µg/disk).
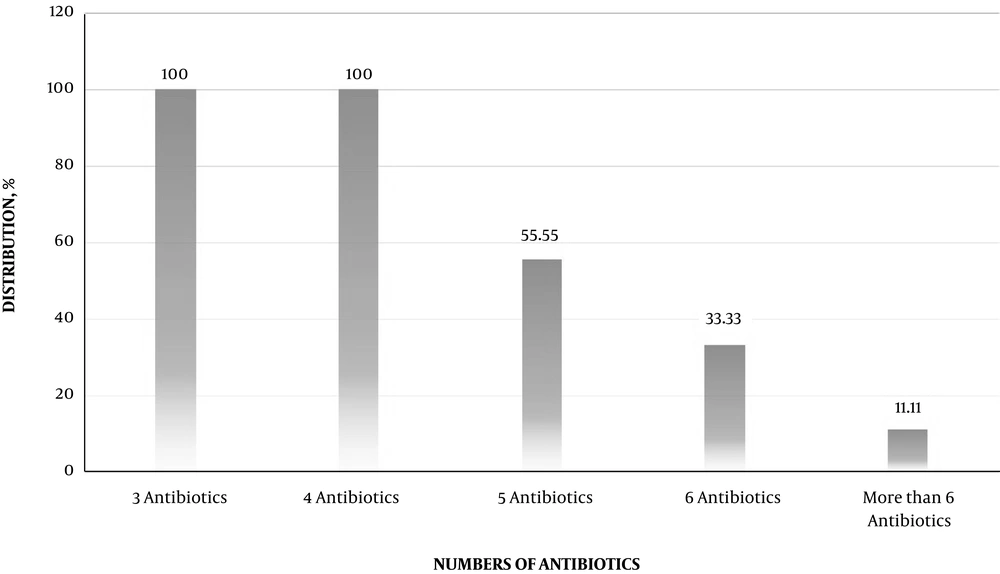
Figure 3 characterizes the distribution of multidrug resistant E. coli O157 bacteria isolated from hospital food samples. Multi-drug resistant E. coli O157 bacteria were determined as those, who had at least simultaneous resistance against three or more than three types of antibiotics. All E. coli O157 bacteria had at least resistance against three types of antibiotics (100%), while distribution of resistance against more than six antibiotic agents was 11.11%.
5. Discussion
Escherichia coli O157 is considered as one of the most dangerous causes of gastrointestinal disorders, along with the cause of most cases of food poisoning around the world (13, 14). Impact of E. coli O157 as a food-borne pathogen is increased in public places, such as dormitories and hostelry centers, especially hospitals. This is because of the presence of weak and immunosuppressed patients in hospitals, which need to use healthy and hygienic food.
The current research was done to evaluate the frequency of virulence genes and phenotypic pattern of antibiotic resistance of E. coli O157 bacteria recovered from hospital food. Apparently, the current examination is the initial description of the molecular characterization and antibiotic resistance properties of E. coli O157 bacteria isolated from gavage and soup samples produced in hospitals. The current research established that 4.5% of hospital food samples were contaminated with E. coli O157 bacteria, which was considerably high. The main reason for the high prevalence of E. coli O157 in hospital food samples is the presence of bacteria in raw meat and chicken used for preparation of soup and gavage samples and then their survival, probably due to the lack of adequate time and also temperature needed for catering and cooking of these ingredients.
The possibility of transmission of E. coli O157 from infected chefs and staffs of the hospital into the food samples is another important probable risk factor for considerable prevalence of E. coli O157 in hospital food samples. Higher distribution of E. coli O157 in gavage samples (6%) compared with soup samples (3%) is due to the fact that preparation of gavage samples requires hand-intervention of staffs of the hospital kitchen. Otherwise, preparation of gavage samples requires high hand manipulation, which increase the risk of microbial contamination. Moreover, liquid and watery basis of gavage samples makes them more prone to become infected with dangerous food-borne bacteria.
Initial investigation, which was conducted in this field, indicated that hospital food was a routine source of E. coli serotypes (15). Ranjbar et al. (1) reported that 39 out of 580 (6.72%) hospital food samples were contaminated with E. coli and distribution of O157 serogroup was 25%. Impact of food handlers in transmission of E. coli bacteria to food has been described before from USA (16) and Nigeria (17). Boost variances in the distribution of E. coli bacteria described in different researches may be due to differences in types of samples, way of sampling, method of experimentation, levels of hygiene, and as a final point topographical and geographical area, from which the food samples were collected.
The current research indicates that E. coli O157 bacteria harbored high distribution of virulence factors and particularly stx1, eaeA, and ehlyA. The EHEC subtype of E. coli and mainly E. coli O157 bacteria should simultaneously harbor stx1, eaeA, and ehlyA genes together. The current findings showed that simultaneous distribution of stx1, eaeA, and ehlyA genes together was 100%. Boost distribution of these factors in E. coli O157 bacteria disclosed their high pathogenicity for patients.
Concurrent presence of virulence genes in some E. coli O157 bacteria specified an imperative public health concern facing health care units and hospitals. Simultaneous presence of virulence genes has also been reported formerly in the E. coli bacteria isolated from diverse kinds of food samples in Iran (1, 3-8, 10, 11), Iraq (18), United States (19), and Nigeria (20). These genes are mostly related to bacterial linkage and attack the cells of gastric epithelium and also occurrence of diarrhea (21). Thus, their boost distribution in food samples of the current study guarantees the high pathogenicity of E. coli O157 bacteria.
This study also suggested that E. coli O157 bacteria displayed high prevalence of resistance against extensive ranges of antibiotics and particularly tetracycline, gentamycin, ampicillin, mezlocillin, enrofloxacin, and trimethoprim. Furthermore, 11.11% of E. coli O157 bacteria harbored simultaneous resistance against more than six antibiotic agents. Unlawful and unselective prescription of antibiotics and inattentiveness to findings of the disk diffusion method in antibiotic prescription are the main factors causing boost prevalence of antibiotic resistance. The current research also established that occurrence of resistance against chloramphenicol in the studied E. coli O157 bacteria was 22.22%. Extraordinary occurrence of resistance against this antibiotic was mostly due to the boost and unequal prescription of chloramphenicol in poultry farms in Iran and transmission of chloramphenicol-resistant E. coli O157 bacteria from contaminated poultry meat to soup and gavage samples.
Another finding of the current study was that the E. coli O157 bacteria had extraordinary prevalence of resistance against animal-based antibiotics, which can indirectly support their animal origin. This part of the research was related to those of India (22) (extraordinary occurrence of resistance against amikacin, erythromycin, kanamycin cephalothin, and gentamicin antibiotics), South Africa (23) (extraordinary occurrence of resistance against gentamicin, ampicillin, and tetracycline antibiotic agents), Korea (24) (extraordinary occurrence of resistance against streptomycin, ampicillin, tetracycline, and amikacin antibiotics), and Mexico (25) (extraordinary occurrence of resistance against trimethoprim-sulfamethoxazole, ampicillin, cephalotine, and chloramphenicol antibiotics). Stewardson et al. (26) described that prevalence of resistance of E. coli bacteria against meropenem, gentamicin, ciprofloxacin, cotrimoxazole, and fosfomycin was 100%, 90%, 87%, 79%, and 98%, respectively.
Extraordinary prevalence of resistance against tetracycline, ampicillin, gentamicin, mezlocillin, enrofloxacin, and trimethoprim was also reported from USA (27), China (28), and Iran (29). Boost prevalence of resistance against routine types of antibiotic agents has been described previously (30-43). Mashak (44) reported that prevalence of E. coli in raw meat, milk, and vegetables collected from Iran was 14%, 20%, and 31.25%, respectively. She showed that all of the E. coli O157 strains harbored simultaneous presence of stx1, eaeA, and ehly virulence genes, which was similar to the current report. She also showed that prevalence of resistance of STEC strains against ampicillin, gentamycin, tetracycline, and ciprofloxacin was 100%, 90.47%, 85.71% and 71.42%, respectively.
In keeping with the high importance of the present research, there were some limitations, including low number of hospital food samples, low variation of hospital food samples, and finally high cost of the study experiment. Resistant food-borne pathogens, such as E. coli O157, methicillin-resistant Staphylococcus aureus (MRSA), Helicobacter pylori, and other bacterial agents are considered as important causes of outbreaks of food poisoning, especially in public places, such as hospitals (45-53). Proper cooking of raw foods, especially meat and chicken, and full compliance of principles of individual hygiene in kitchen of hospitals are the best ways to increase microbial quality and safety of hospital food samples, especially gavage and soup.
5.1. Conclusions
In conclusion, the current study recognized a high distribution of virulence genes and antibiotic resistance in E. coli O157 bacteria recovered from gavage and soup hospital foods. Gavage samples had a higher incidence of E. coli O157 bacteria. Stx1, eaeA, and ehlyA virulence factors, resistance against ampicillin, gentamycin, and tetracycline and presence of multi-drug resistant bacteria were the most frequent characters in the E. coli O157 bacteria recovered from soup and gavage. Simultaneous presence of stx1, stx2, eaeA, and ehlyA virulence genes exhibits the boost pathogenicity of E. coli O157 bacteria. Presence of O157 bacteria, virulence factors, and animal-based antibiotics in soup and gavage posed inadequacy of cooking temperature and time used for preparation of foods in hospitals. It appears that there were no severe managements on the principles of food security in hospitals. Attention to the findings of disk diffusion method, and whole cooking of foods can diminish the risk of E. coli O157 bacteria in soup and gavage samples produced at hospitals.