1. Background
The genus Burkholderia is characterized by taxonomic complexity and includes such highly pathogenic species as Burkholderia pseudomallei and B. mallei, the causative agents of melioidosis and glanders, respectively, and opportunistic pathogens of B. cepacia complex. Burkholderia cepacia complex as well as B. pseudomallei are highly resistant to antimicrobials, however, they have different resistance patterns (1). Therefore, the correct and well-timed identification of these pathogens is urgent for successful treatment. Furthermore, the wrong identification of highly pathogenic bacteria increases the risks for laboratory stuff due to potential unsafe manipulation. The currently available biochemical systems widely used in clinical microbiology laboratories can misidentify some Burkholderia species. The certain problem is associated with the misidentification of B. pseudomallei as B. cepacia and vice versa using Vitek 2 platform (bioMerieux, France) (2, 3).
2. Objectives
This work aimed to clarify the results of identification of clinical Burkholderia spp. isolate using species-specific PCR and MLST profiling of whole-genome sequence data.
3. Methods
3.1. Case Description
A case of septicemia with severe fever in a 68-year-old woman was preliminarily diagnosed as ‘melioidosis’ in Phu Tho Provincial General Hospital in January 2018. In anamnesis, there were no risk factors such as diabetes, AIDS, cystic fibrosis, or immune system diseases.
3.2. Isolation and Identification of the Strain
A non-fermenting Gram-negative rod was isolated from the blood of a patient. The strain was subcultured on LB agar (HiMedia Laboratories Pvt. Ltd, India) for 24 hours at 37°C before testing with the Vitek 2 GN cards (bioMerieux, France), according to the manufacturer’s instructions. The antibiotic susceptibility was tested according to CLSI recommendation (4).
3.3. Molecular Methods
The multi-locus PCR assay targeting an individual set of chromosomal beta-lactamase genes in B. pseudomallei and B. cepacia complex was performed as described previously (5). Genomic DNA of the strain was isolated using GeneJET genomic DNA purification kit (Thermo Scientifics, Lithuania). The pair-end library was constructed via Nextera XT library preparation kit (Illumina Inc., USA). Sequencing was performed on the Illumina MiSeq platform (Illumina Inc., USA).
3.4. Bioinformatics Tool
Filtered reads were assembled de novo using SPAdes 3.11 software (6). The sequences of seven house-keeping genes, according to the MLST scheme by Spilker et al. (7), were matched in assembly using the BLASTn algorithm. Multiple alignments and phylogenetic analysis were performed using the MEGA 7 package (8). Identification of related genotypes in the MLST dataset was performed using eBURST v3 algorithm (http://eburst.mlst.net/).
4. Results
The isolated strain was identified initially as B. pseudomallei using Vitek 2 GN (bioMerieux, France) with a reliability of 87%. The strain was resistant to gentamicin, colistin, and amoxicillin/clavulanate. It was also intermediate resistant to imipenem and was sensitive to ceftazidime, meropenem, doxycycline, and co-trimoxazole. The observed amoxicillin/clavulanate resistance pattern and colony morphology on Ashdown agar cast doubt on the results of biochemical identification. The strain was additionally tested using a recently developed multiplex PCR for detection of an individual set of chromosomal class B and D beta-lactamase genes in B. pseudomallei and B. cepacia complex (5). The analysis suggested that the isolate belongs to B. cepacia. The strain was designated as B. cepacia PT02. Three months later of initial biochemical testing, the strain was identified as B. cepacia by Vitek 2 with a reliability of 91%.
Whole-genome sequencing of B. cepacia PT02 was carried out for final confirmation of obtained results. The obtained contigs were deposited in the Genbank database (accession no. QLUZ00000000). The established MLST profile of the isolate was added to PubMLST B. cepacia complex database (https://pubmlst.org/bcc) with ID 2680. The MLST analysis revealed new variants of gltB and gyrB alleles, which were not previously described. The resulting allelic profile of B. cepacia PT02 was assigned to ST1400, which was not presented at the time in Pub MLST. The rMLST profiling of PT02 revealed the concordance in 42 out of 53 loci used in the database.
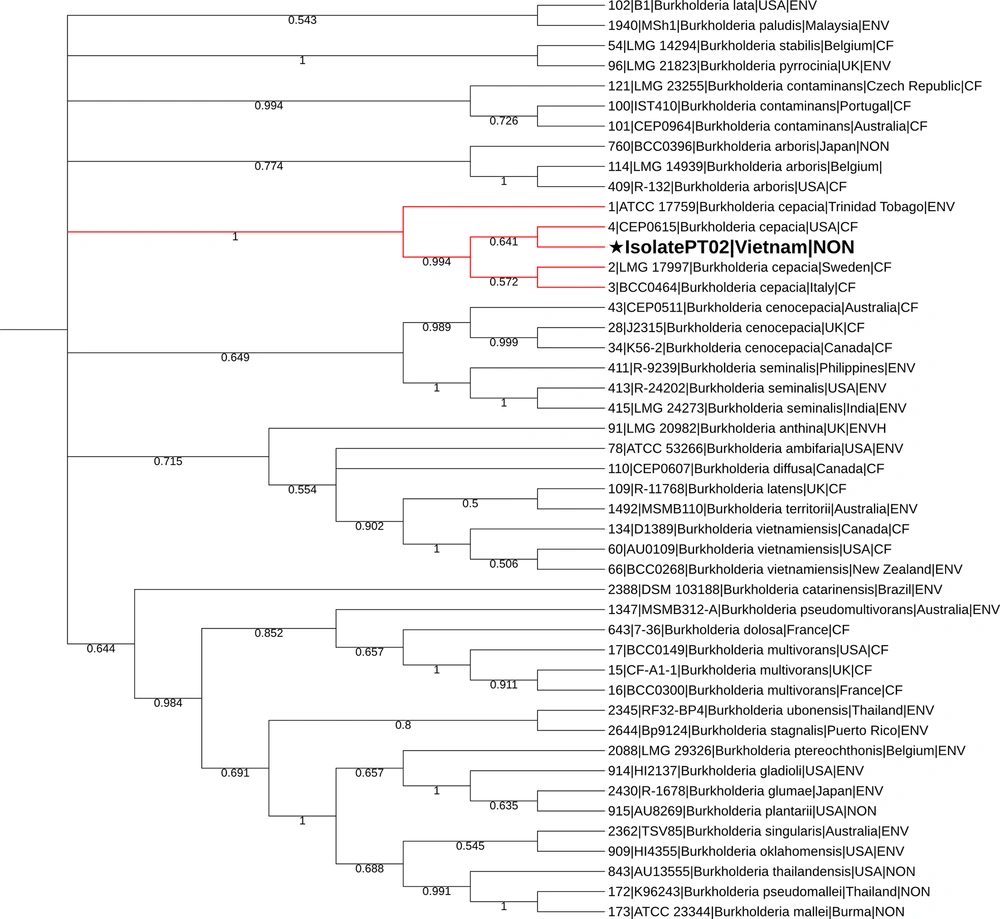
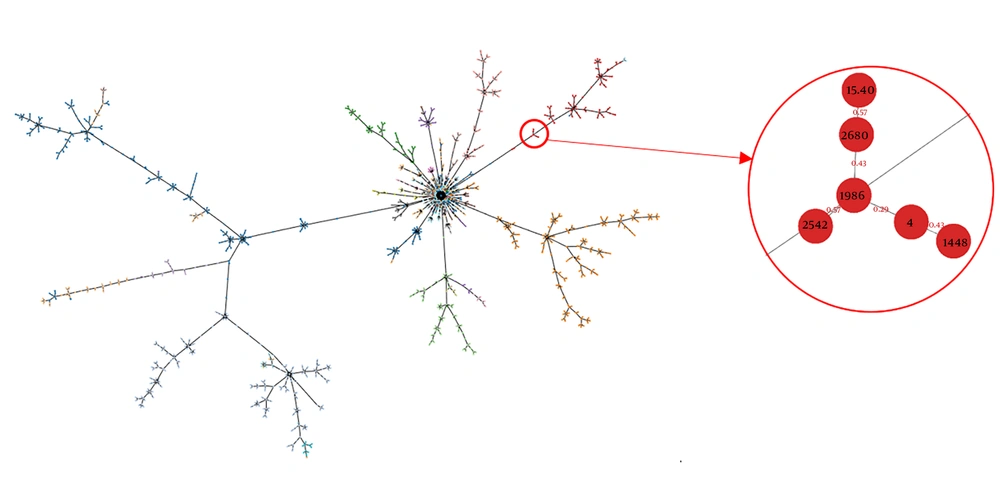
According to Maximum Likelihood phylogeny (bootstrap = 1000), the PT02 isolate belongs to the B. cepacia clade of the tree (Figure 1). The similar results were obtained by eBURST analysis of allelic profiles of PT02 and 2688 B. cepacia complex strains from the MLST database. The PT02 isolate also belonged to B. cepacia branch and joined the clonal complex of B. cepacia CVS136 (Figure 2). The root strain CVS136 (Taiwan) and strain R-66653 (Belgium) were most closely related to PT02 (the genetic distance of 0.43 and 0.57, respectively). It should be noticed that all strains of the CVS136 clonal complex were isolated from patients or hospital environment. Thus, B. cepacia R-52513 and R-66653 were isolated from non-cystic fibrosis patient (NON) in Belgium, B. cepacia CEP0615-from cystic fibrosis patient in the USA (CF), and B. cepacia NML171677-from non-cystic fibrosis patient with septicemia in Canada (NON, blood culture).
The genetic distance of 45 concatenated sequences of Burkholderia spp., and B. cepacia PT02 house-keeping genes according to MLST scheme (7). Maximum Likelihood dendrogram (bootstrap = 1000) constructed using the TamuraNei model. The consensus bootstrap tree was obtained by 1000 iterations to demonstrate the reliability of clade forming. The multiple alignments were performed using MUSCLE algorithm. Alignment blocks containing gaps and missing data were excluded from analysis. The final dataset contained 2,271 sites.
Population snapshot of 2688 Burkholderia spp., constructed with the eBURST algorithm according to allelic profiles obtained from the PubMLST Bcc database. The red circle highlights the clonal complex of B. cepacia CVS136 (node 1986) including isolate PT02 (node 2680). The relative distance between strains indicated above the rib of the graph.
5. Discussion
The significant phenotypic variability is common in B. cepacia and other B. cepacia complex species. In addition, there has been controversy regarding the optimal differentiation system for B. pseudomallei complex and B. cepacia complex species. Although conventional tests (colony morphology on selective media, antimicrobial resistance patterns, agglutination) usually give acceptable discrimination, they are time-consuming. The misidentification of B. pseudomallei as B. cepacia and vice versa by commonly used Vitek 2 system was repeatedly described (3, 9, 10). Recently, Podin et al. noted that two enzymatic tests, NAGA (beta-N-acetyl-galactosaminidase) and BNAG (beta-N-acetyl-glucosaminidase), were distinct between correctly and misidentified B. pseudomallei isolates (11). In addition, the combinations of positive dCEL (D-cellobiose) and negative ProA (L-proline arylamidase), TyrA (tyrosine arylamidase), or NAGA were observed in B. pseudomallei strains incorrectly identified as B. cepacia (12).
The B. cepacia PT02 described here was initially positive for dCEL and TyrA, which leads to misidentification of the isolate as B. pseudomallei on Vitek 2 platform. Subsequently, the negative TyrA test, after a three-month storage period, was a key biochemical feature to identify the isolate as B. cepacia. The metabolic plasticity and variable enzymatic profile complicate the reliable biochemical differentiation within B. cepacia complex species, as well as from other Burkholderia spp. (13). Currently, this is the main reason why the molecular methods have a preferential use for B. cepacia complex differentiation. To date, there is a wide range of PCR-based methods developed for rapid identification of B. cepacia complex species (14, 15) and their differentiation from B. pseudomallei complex (5, 16, 17). The described detection of a B. cepacia complex specific set of beta-lactamase genes and sequence type determination allowed us to confirm the studied isolate decisively as B. cepacia.
5.1. Conclusions
The complex application of various molecular methods, considering their growing availability and high discriminatory power, in some cases can provide the most accurate and rapid identification of pathogenic Burkholderia, and thus, reduce risks of improper treatment or biosafety threat. The data mentioned above also demonstrates a better resolution of high-throughput sequencing/in silico MLST vs. conventional B. cepacia complex genomovar determination by recA restriction fragment length polymorphism.