1. Background
Dental caries is a transmissible infectious and multifactorial disease associated with the mutans streptococci group. The group includes seven species, with Streptococcus mutans as the best-known species. The acquisition of mutans streptococci in children occurs during the first year of life, which is called the window of infectivity (1). The principal source from where infants acquire mutans streptococci is thought to be their main care provider, especially the mother, because the mothers and children present similar or identical bacterial profiles (2, 3). Some authors have suggested that mutans streptococci acquisition in children increases with the erupted teeth (4). Streptococcus mutans is considered the primary species closely associated with the etiology of dental caries in humans, due to its ability to produce acid and extracellular polysaccharides (5, 6). Recent studies suggest that there is a wide genetic diversity in S. mutans strains associated with caries in children (7-11).
Many methods have been used to identify the mutans streptococci (MS) diversity, such as DNA genomic fingerprinting by pulse-field gel electrophoresis (PFGE) (12), arbitrary-primer polymerase chain reaction (AP-PCR) (11), repetitive extragenic palindromic PCR (rep-PCR), and multilocus sequence typing (MLST) (13). PCR based upon the dexA gene sequence has been successfully applied for the identification of S. mutans species. RFLP analysis of the PCR-amplified 16S rRNA gene has also been used for the identification of MS species (14-16). However, the 16S rRNA gene PCR-RFLP analysis has not been applied yet to differentiate the genotypes of S. mutans species. On the other hand, strains of S. mutans that are exclusively cariogenic have not been genotyped, diversity has always been observed (7, 11); the genetic stability of these strains has neither been studied extensively. We only found a prospective study on the diversity and genetic stability of the strains of cariogenic S. mutans (8).
2. Objectives
The aim of this study was to identify the genotypic diversity of S. mutans types and their association with caries in schoolchildren through 16S rRNA gene PCR-RFLP analysis of S. mutans isolates in two episodes.
3. Methods
3.1. Subjects
In 2013, we initiated the study in 7 - 8-year-old schoolchildren from a state-funded elementary school in Mexico City. Table salt available in Mexico City is all fluoridated. In this area, the fluoride content in water was < 0.247 ppm. We registered 36 children in the second school year. The subjects’ families belonged to the mid socioeconomic status. All the children were from the same grade and staying at the school for eight hours per day. They had breakfast and lunch at the school canteen and performed unsupervised tooth brushing after every meal. These children had three holiday periods: 15 days of spring break, 10 days for year-end holidays, and 45 days between grades, during which their diet was not regulated. None of the schoolchildren had chronic diseases and received no antibiotic therapy and fluoride therapy during the last two weeks before biofilm sampling. None of them used orthodontic appliances. Oral samples were collected simultaneously with the clinical examination (baseline sample), and the procedure was repeated 12 months later.
3.2. Clinical Examination
Two calibrated examiners (Kappa = 0.95) performed the oral examination for caries prevalence at baseline using a natural light source, a mirror, and a periodontal probe. The caries prevalence was scored by the sum of decayed, missing, and filled deciduous surfaces (dmfs) and decayed, missing, and filled permanent surfaces (DMFS), according to the World Health Organization criteria (17). To determine the caries incidence, the oral examination was repeated 12 months later by the same examiners. To avoid bias, the schoolchildren were evaluated without access to their caries record. For each child, the caries increment was calculated by subtracting the baseline dmfs or DMFS score from the new dmfs or DMFS score.
3.3. Biofilm Sampling
Two hours after the most recent meal, the bacterial dental biofilm was obtained from the right lower first permanent molars by scraping 10 times from distal to mesial central fissures and pits using a short sterile hypodermic needle (18). The needle was then immersed in 2 mL of trypticase soy broth (TSB) (Difco/Becton Dickison, Sparks, MD, USA), supplemented with 200 U/L of bacitracin and 20% sucrose, using three sterile glass beads. Biofilm samples were stored on ice while transported to the laboratory and processed within two hours of collection. The samples were vortexed (Thermo Fisher Scientific, Inc., USA) for 30 s and used undiluted for further analysis.
3.4. Streptococcus mutans Counts
Biofilm samples were inoculated on selective mitis salivarius agar (Difco/Becton Dickison, Sparks, MD, USA) containing 200 U/L of bacitracin and 20% sucrose (19). Culture plates were incubated in 5% CO2 at 37ºC for 72 h. After incubation, characteristic S. mutans colonies were counted and the number of colony-forming units per milliliter (CFU/mL) was determined. To present the data, the counts were transformed into an ordinal scale of < 104 to 106 and analyzed.
3.5. Streptococcus mutans Identification
One colony resembling S. mutans (rough appearance and adherent to the agar) from each sample was transferred to 8 mL of TSB containing 200 U/L of bacitracin and 20% sucrose, and incubated at 37°C for 48 h. The genomic DNA of the isolates cultured in TSB was extracted using the Wizard® Genomic DNA Purification kit (Promega Corporation, Madison, WI, USA) according to the instructions provided by the manufacturer with a modified lytic enzymes mixture consisting of 10 mg/mL lysozyme and 10 mg/mL mutanolysin (20). Prior to genotyping by PCR-RFLP analysis, all isolates were confirmed as S. mutans using S. mutans-specific primers for the dexA gene to specifically amplify a 1272-bp fragment, based on the description provided by Igarashi et al. (14, 21). The dexA gene was amplified with the primers, SD1, 5´-TAT GCT GCT ATT GGA GGT TC-3´ and SD2, 5´-AAG GTT GAG CAA TTG AAT CG-3´. The PCR mixture consisted of 1X PCR buffer, 1.5 mM MgCl2, 60 ng/µL acetylated bovine serum albumin, 200 µM each of dNTPs, 0.5 µM of each primer, 2.5 U of Taq DNA polymerase (Promega Co.), and 1.0 µg of DNA. Amplification was performed in a thermocycler (My Cycle®, Bio-Rad Laboratories, Hercules, CA, USA) according to the conditions previously described by Igarashi et al. (14, 21) with some modifications: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, an extension at 72°C for 1 min, and the final extension at 72°C for 10 min.
3.6. Streptococcus mutans Genotyping
Streptococcus mutans genotyping was performed by 16S rRNA gene PCR-RFLP analysis using the universal primers 8F and 1492R. Amplification was performed according to the conditions previously described by Sato et al. (16). The primer sequences were 8F, 5´-AGA GTT TGA TCC TGG CTC AG-3´ and 1492R, 5´-TAC GGG TAC CTT GTT ACG ACT T-3´. The PCR mixture consisted of 1X PCR buffer, 3.0 mM MgCl2, 200 µM each of dNTPs, 1.0 µM of each primer, 2.5 U of Taq DNA polymerase, and 1.0 µg of DNA. Amplification was performed by: initial denaturation at 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 40 s, an extension at 72°C for 40 s, and the final extension at 72°C for 7 min.
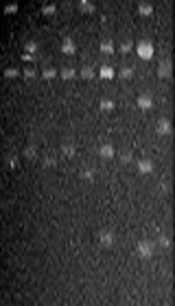
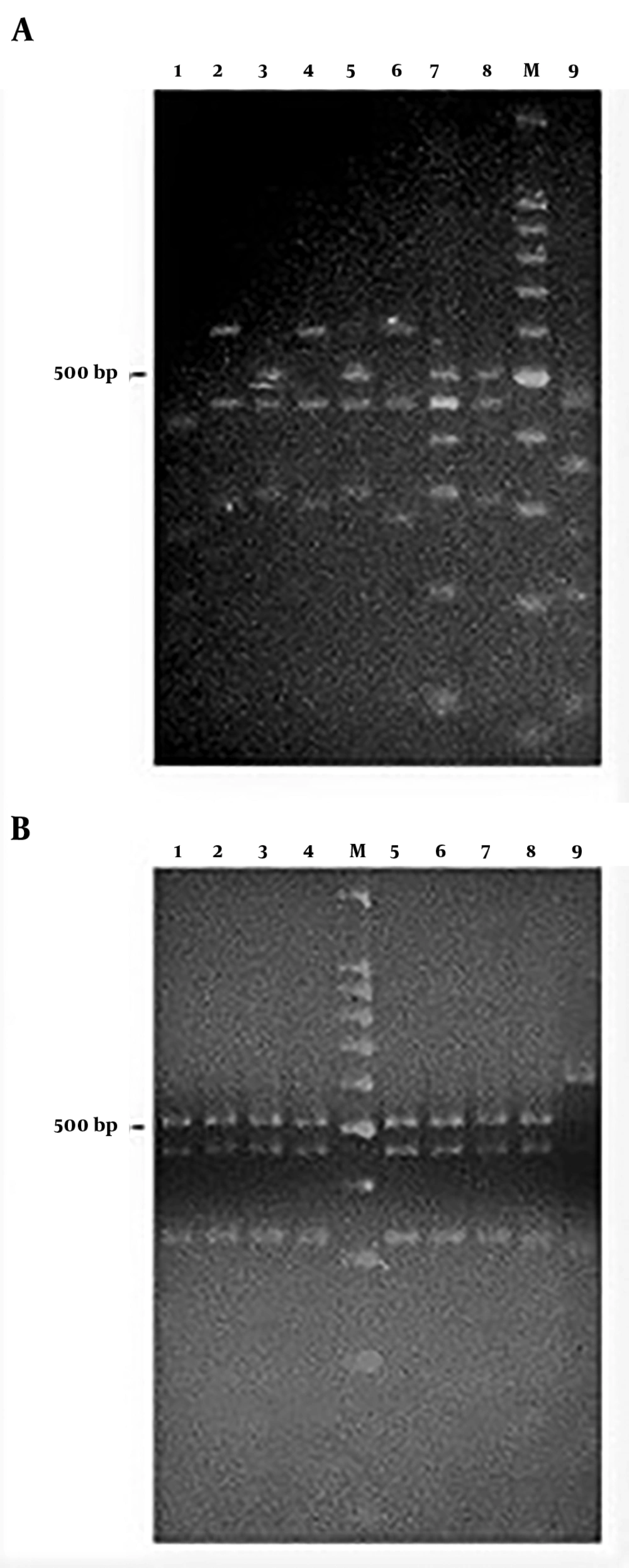
The amplicons of the 16S rRNA gene were digested with restriction endonuclease HaeIII (Promega Co.). The digestion mixture consisted of 1X buffer C, 10 µg/µL acetylated bovine serum albumin, HaeIII 10 U/µL, and 1 µg of PCR product. Digests were incubated at 37°C for 90 min and separated on a 3.0% agarose gel in Tris-borate EDTA buffer at 80 V for 5 h. Images were captured with the Gel-Doc™ XR System (Bio-Rad, Hercules, CA, USA). The products’ digestion patterns were analyzed to create dendrograms with Gene Tools and Gene Directory software (Syngene, Cambridge, UK). Similarity percentages were obtained from the unweighted pair group with mathematical average (UPGMA) based on Dice coefficients. Band position tolerance was set at 1.25%, which provided a hierarchical cluster representation of similarities between samples and indicated S. mutans strain level grouping. A similarity coefficient of > 95% was considered to define the genotype.
3.7. Statistical Analysis
Statistical analysis was performed using the JMP® version 8.0.2 program (SAS Institute Inc., NC, USA). The chi-square test and Fisher’s exact test were applied to determine differences in the distribution of S. mutans genotypes according to gender at baseline and after one year, respectively. The Student’s t-test was used to evaluate differences in dmfs/DMFS scores and S. mutans counts of all the children between baseline examination and one-year examination. The analysis of variance (ANOVA) and Student’s t-test were applied to evaluate differences in dmfs/DMFS values and S. mutans counts between genotypes at baseline and after one year, respectively. Values of P < 0.05 were considered significant.
4. Results
For the identification of S. mutans carriers, from the 36 schoolchildren, 11 (30%) dropped out after one year and were not included in the analysis. We found 25 S. mutans isolates confirmed by dexA-PCR in 14 boys and 11 girls. The children's age was 7.2 ± 0.4 years. Streptococcus mutans biofilm counts were determined in all children. At baseline, eight children (32%) presented S. mutans counts of lower than 104 CFU/mL, eleven (44%) showed counts between 104 and 105 CFU/mL, three (12%) exhibited counts between 105 and 106 CFU/mL, and three (12%) had counts of higher than 106 CFU/mL. After one year, 17 children (68%) showed S. mutans counts of lower than 104 CFU/mL, four (16%) presented counts between 104 and 105 CFU/mL, and four (16%) presented counts between 105 and 106 CFU/mL. The mean of S. mutans among the 25 study children was 7.0 × 105 ± 2.0 × 105 UFC/mL at baseline and 4.8 × 104 ± 1.0 × 104 UFC/mL after one year without statistically significant differences (P = 0.12) (Table 1).
| Genotype | P Value | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| Baseline | ||||||
| Number of children | 13 | 9 | 1 | 1 | 1 | |
| Caries index | ||||||
| dmfs | 5.0 ± 5.9 | 3.4 ± 4.9 | 0 | 0 | 0 | 0.7227 |
| DMFS | 0.5 ± 1.4 | 0.2 ± 0.7 | 0 | 0 | 0 | 0.9729 |
| Streptococcus mutans countsb | ||||||
| CFU/mL | 7.3 × 105 ± 2.0 × 105 | 4.1 × 104 ± 6.5 × 104 | 7.4 × 106 | < 104 | 1.2 × 105 | 0.0063 |
| After Twelve Months | ||||||
| Number of children | 23 | 2 | 0 | 0 | 0 | |
| Caries index | ||||||
| dmfs | 3.2 ± 4.9 | 2.0 ± 2.8 | - | - | - | 0.7404 |
| DMFS | 0.6 ± 1.5 | - | - | - | - | 0.5863 |
| Caries incidence in both dentitions | 1.2 ± 1.8 | 1.3 ± 2.2 | - | - | - | 0.9746 |
| S. mutans countsb | ||||||
| CFU/mL | 5.1 × 104 ± 1.0 × 104 | 1.4 x 104 ± 1.9 x 104 | - | - | - | 0.6331 |
zAbbreviations: dmfs, decayed, missing or filled deciduous surfaces; DMFS, decayed, missing or filled permanent surfaces.
aValues are expressed as mean ± SD.
b Mean CFU of S. mutans counts among the study subjects.
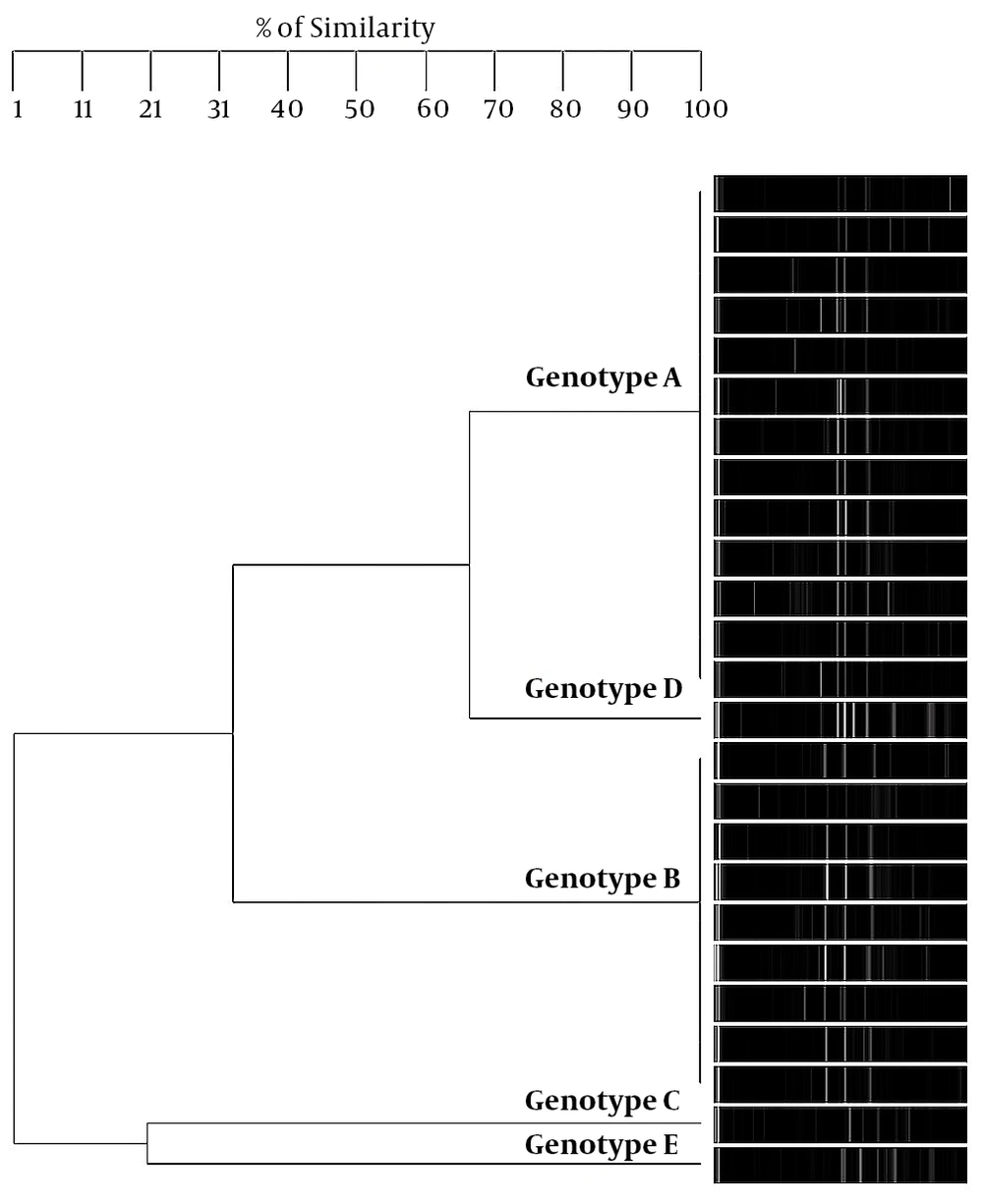
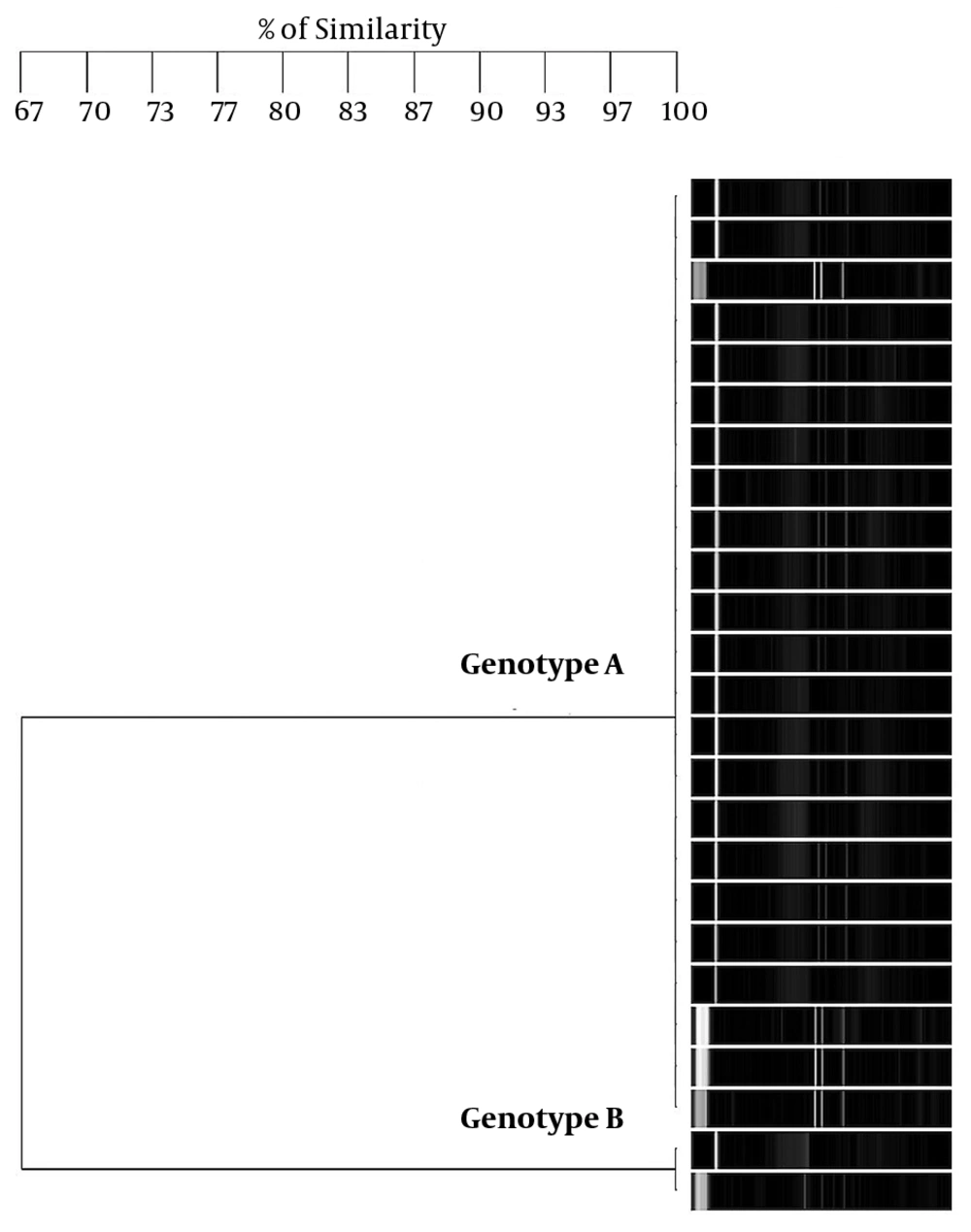
Regarding S. mutans genotyping at baseline, five genotypes were found among the 25 isolates (Figure 1A). Genotype A (52%) and genotype B (36%) were more frequent. C, D, and E genotypes were found in one child only (Figure 2). After 12 months, the genotypic diversity was reduced from five to two (C, D, and E genotypes were not found) (Figure 1B). In this episode, 23 children presented genotype A (92%) and genotype B was identified in only two children (Figure 3). There was no difference between genders in the distribution of genotypes at baseline (P = 0.5799) and after 12 months (P = 0.4867). Of the 14 boys (56%), seven presented genotype A, five genotype B, one genotype C, and another genotype E. Of the 11 girls, six presented genotype A, four genotype B, and one genotype D. No significant differences were found in relation to the distribution of genotypes by gender and age (P = 0.5799). When analyzing the genotype by age, giving weight to the gender variable, age did not influence the distribution of genotypes (P = 0.4177). Therefore, the data are presented for the whole study group.
PCR-amplified 16S rRNA genes from Streptococcus mutans strains isolated from the schoolchildren and digested with HaeIII at baseline (A) and after one year (B). (A) Lane: 1, genotype C; lanes: 2, 4, and 6, genotype B; lanes: 3, 5, and 8, genotype A; lane: 7, genotype D; lane: 9, genotype E. (B) Lanes: 1-8, genotype A; lane: 9, genotype B. M, Molecular size markers (100 bp DNA ladder, Promega Co.).
Regarding the caries prevalence at baseline, the 25 children had a mean dmfs of 3.84 ± 5.28 and a mean DMFS of 0.32 ± 1.06. No significant differences were found between dmfs/DMFS scores at baseline and after one year (P = 0.62). The dmfs/DMFS scores were not statistically associated with the genotypes at baseline and after 12 months. However, the children with genotype A presented more caries lesions than children with other genotypes (Table 1). The analysis of variance revealed that those children harboring genotypes A and C had the highest S. mutans counts at baseline (P = 0.0063). In contrast, no associations were found between S. mutans counts and genotypes A and B after 12 months (Table 1). The Student’s t-test revealed that baseline and final S. mutans counts did not differ among genotype A carriers (P = 0.1169).
5. Discussion
In this study, 16S rRNA gene PCR-RFLP analyses were used to evaluate the genotypic diversity and stability of S. mutans strains isolated from a group of schoolchildren. These analyses have been successfully applied for the differentiation of mutans streptococci species (16). However, this analysis had not been used to genotype S. mutans strains. For the genotyping of S. mutans, the techniques of AP-PCR (11), rep-PCR, and MLST (13) have been used so far. However, these techniques are usually more laborious and expensive, so the use of the 16S rRNA gene PCR-RFLP analysis offers a simpler and cheaper method to compare genotypic diversity and stability of S. mutans. The results of this study showed that S. mutans displayed a genetic diversity in children with caries, which is in line with previous reports (7, 9-11, 22). With respect to the group of schoolchildren studied in this work, the genotypic diversity of S. mutans included five genotypes at baseline, two of which were more frequent: genotype A and genotype B (Table 1).
Interestingly, we noticed that children who carried genotype A, which was more prevalent at baseline and 12-month follow-up than the other found genotypes, showed higher values of CFU/mL, as well as the highest dmfs and DMFS scores (Table 1). When running a model giving weight to the number of CFU/mL, into which caries indices were entered as the dependent variables and persistence of genotype as the independent variable, we found that genotype A was influenced by the amount of CFU/mL. These results show that S. mutans counts and dmfs/DMFS indices were higher in those children harboring genotype A, except the child who presented genotype C. In this sense, a positive correlation has been reported between S. mutans counts in biofilm and caries indices, showing that children with high counts of S. mutans had high caries scores (23, 24).
After 12 months, a loss of genotypic diversity of S. mutans was observed, with genotype A still as the dominant genotype (Figure 3). To the best of our knowledge, only one study has investigated.
The stability of genotypes over time (8), reporting that genotypes decreased and only prevailed the dominant genotypes. We postulate that genotype A possibly colonized children more easily and displaced other genotypes after one year. Thus, the presence of cariogenic competing bacteria with probably different virulence abilities, under favorable environmental conditions, may facilitate the occurrence of microevolution and might influence the stability and adaptability of certain S. mutans genotypes in the oral cavity. These changes in the microenvironment and the stability of strains can also be observed in the findings of Dame-Teixeira et al. (22), in which, despite decreasing the number of genotypes after partial dentin caries removal, the virulence of the strains remained.
The reduction in the number of genotypes observed after one year could be associated with the horizontal transmission of genotype A as a predominant genotype of S. mutans among the schoolchildren in this study. This assumption might be influenced by the fact that our study group had breakfast and lunch at school that is in the same place, which may promote horizontal transmission. Additionally, as mentioned above, children brushed their teeth after every meal without supervision. In this regard, it has been reported that cultural differences, such as eating with the same cutlery, sharing the same toothbrush, or defective washing up routines, increase the possibility of horizontal transmission (25-27). The variability in transmission can be associated with individual susceptibilities (1).
5.1. Conclusions
In conclusion, we found diversity in the S. mutans genotypes among the study group of schoolchildren. This diversity of genotypes decreased after 12 months and the more stable genotype associated with dental caries predominated.