1. Background
Periapical pulp disease is a common oral disease. Currently, the most effective treatment for periapical pulp disease is root canal treatment (1). Despite the continuous development of treatment technology, the success rate of the first root canal therapy is only 85% to 90%, and the success rate of the second root canal treatment is even lower (2). Most treatment failures are caused by Enterococcus faecalis (2).
Enterococcus faecalis are Gram-positive bacteria that can appear singly, in pairs, or short chains (3). Enterococcus faecalis survive very hard environments, such as high salt concentrations and extreme pH (4). Enterococcus faecalis are common bacteria in the oral cavity. The content of E. faecalis in failed root canal treatment cases is nine times more than that of primary endodontic infections (5). Enterococcus faecalis can suppress the activity of lymphocytes contributing to the endodontic failure (6). Enterococcus faecalis causes diseases mainly depending on its survival ability and pathogenicity in the root canals of teeth. It conquers the survival challenges in several ways. It binds to dentin by possessing serine protease, gelatinase, and collagen-binding protein. It invades and lives in dentinal tubules.
Biofilms offer several benefits of E. faecalis (7). Enterococcus faecalis residing in a biofilm grow much more slowly than the others do. As a result, E. faecalis take up antimicrobials, antibodies, and phagocytosis much more slowly (8). The formation of biofilms is a key toxic factor for many microorganisms causing chronic infections. Multiple factors regulate the biofilm development. The life cycle of biofilm includes four stages: Initial adhesion, early development, mature biofilms, and late-stage dispersal (9). Intracellular second messenger cyclic dimeric (3’→5’) GMP (c-di-GMP) increases biofilm formation and plays a key role in the life cycle of biofilms (3).
The c-di-GMP, a small molecule was first found as an allosteric activator of bacterial cellulose synthase in 1987 (10). During the past 30 years, c-di-GMP has been proved an important and common bacterial second messenger. It regulates cell differentiation, biofilm formation, virulence, and other processes (11, 12). The c-di-GMP binds and affects the activity of effectors such as PilZ, FleQ, PelE, PopA, etc. These c-di-GMP-binding effectors directly control various target processes (13, 14).
The c-di-GMP is synthesized by diguanylate cyclase (DGC) consisting of a GGDEF domain and degraded by phosphodiesterase (PDE) containing either a conserved EAL domain or HD-GYP domain (15). The c-di-GMP is produced from two GTP molecules by DGC. DGC is made up with two subunits with GGDEF domains; each subunit binds one molecule of GTP (16). GGDEF domains act as c-di-GMP cyclase, and mediate the phenotypic changes in some pathogenic organisms (4). Mutations in the GGDEF domain result in the loss of biofilm forming activity in Vibrio cholera (5).
2. Objectives
The current study aimed at identifying gene encoding GGDEF domain and investigating its function in E. faecalis.
3. Methods
3.1. Ethics Statement
The study protocol was approved by the Ethics Committee of the University of Science and Technology of China (0000000A-1). The study was conducted according to the Declaration of Helsinki and current ethical guidelines (2013).
3.2. Analyzing the GGDEF Domain Proteins in Enterococcus faecalis by Bioinformatics
The whole genome sequence of E. faecalis was analyzed through blast and bioinformatics software (NCBI: https://www.ncbi.nlm.nih.gov/). The possible target protein encodes a credible GGDEF domain.
3.3. Bacterial Strains and DR75-RS11090 Plasmid
The E. faecalis were cultured in brain-heart-infusion (BHI) broth (TAKARA, Dalian, China) at 37°C (80% N2, 10% CO2, 10% H2). Flanking regions of DR75-RS11090 open reading frames were amplified by PCR. The primers were: UP-F, atgcggatccAATGGCTGCTATTGGCGGTAA; DW-R, atgcggatccAATGTGTCCGCTGTTTTGGC (Invitrogen, Suzhou, China). PCR products were gel purified and cloned into pUC57 (TAKARA, Dalian, China); the constructed plasmids were named pUC57-UP-amp-DW. The plasmids were then electrically transfected into E. faecalis strains (ATCC, Beijing, China) to construct DR75_RS11090-/- strain. To construct DR75_RS11090+/+, full-length genes were amplified by PCR. The primers were: F, acgcgaattcATGCAAAAGAAGAGAATTCA; R, acgcggatccTCACTCCTGTTCATACATTTC (Invitrogen, Suzhou, China). The amplified products were cloned into pET-28a (+) (TAKARA, Dalian, China)to obtain recombination plasmid pET-28a-DR75_RS11090. After correct sequencing, the constructs were transfected into E. faecalis strains.
3.4. Bacterial Growth and Biofilm Formation
Growth curves and biofilm formation for wild-type, DR75-RS11090+/+, and DR75_RS11090-/- were measured in an automatic microplate reader (Molecular Devices, California, America) (6). The growth curve was repeated five independent times. Biofilm formation was repeated in two independent experiments.
3.5. Transmission Electron Microscopy of Enterococcus faecalis Strains
Enterococcus faecalis strains were grown for 16 hours at 37°C in BHI. Bacterial cells were collected and resuspended in BHI. The bacterial cells were co-cultured with glass slide in 24-well plates. The strain was fixed with 2% glutaraldehyde and scanned under the transmission electron microscopy (TESCAN, Brno, Czech Republic) (7).
3.6. Measurement of the c-di-GMP Acid Production and Acid Resistance
Enterococcus faecalis strains were harvested by centrifugation and resuspended in 1 mL phosphate buffered saline (PBS; TAKARA, Dalian, China). The bacterial cells were collected for analysis by liquid chromatography combined with mass spectrometry (Agilent, Shanghai, China). Enterococcus faecalis strains were grown for 16 hours at 37°C in BHI. Bacterial cells were collected by centrifugation and resuspended in PBS. BHI was prepared at different pH values containing 1% glucose. The bacterial suspension and BHI were mixed with 1:10 (V/V) ratio, anaerobically incubated for 24 hours at 37°C; each test was performed in triplicate. The supernatant was collected after centrifugation. The pH value was measured by the digital acidometer (Jiangdong, Suzhou, China).
Enterococcus faecalis strains were inoculated into Todd Hewitt broth containing 0.3% yeast extract (THYE). The overnight culture was mixed with freshly prepared THYE (pH 7.5) with 1:10 (V/V) ratio, and anaerobically incubated at 37°C for 18 hours. The bacterial cells were collected after centrifugation, resuspended in THYE, and cultured for two hours at 37°C. Before and after incubation, 1 μL of bacterial solution was diluted 1:1000 and cultured on plates, counting the number of bacteria. According to the following formula (8): Acid resistance = number of viable bacteria (pH 5) after incubation/number of viable bacteria (pH 5) before incubation x 100%.
3.7. Water-Insoluble Polysaccharide Production Test
Overnight cultures of E. faecalis were collected and resuspended in sterile water to an OD600 of 1.0. The bacterial cells were mixed with BHI with 1:10 (V/V) ratio, and anaerobically incubated at 37°C for 48 hours. The cells were washed twice with distilled water equal to the volume of the culture medium. After washing, the cells were washed with 0.5 M/L of NaOH (Sangon, Shanghai, China) in equal volumes three times, and the supernatant was collected as water-insoluble extracellular polysaccharide. Three times of the volume of anhydrous ethanol was added to 1.5 mL of supernatant, and incubated at 4°C overnight. The water phase was discarded after centrifugation, dissolving the precipitation in distilled water and 0.1 M/L of NaOH. The content of water soluble and water insoluble glucan were determined by anthrone method (17), tested in OD 620.
3.8. Statistical Analyses
The student t-test was used to determine the statistical significance of the differences. Significant differences were observed in various phenotypes among different strains (P < 0.05) using SPSS.
4. Results
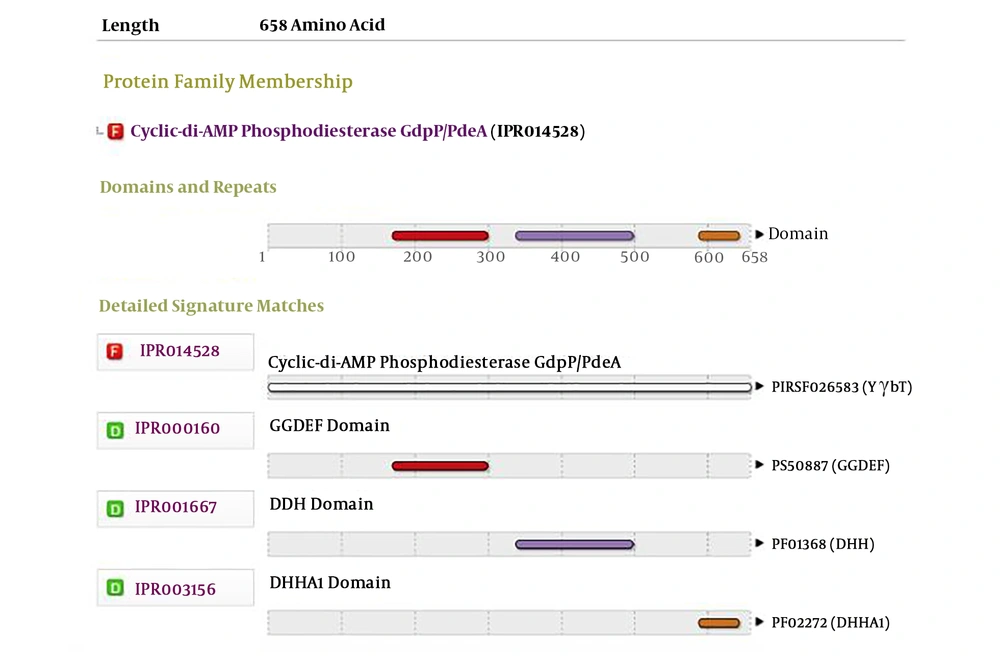
4.1. Analyzing the GGDEF Domain Proteins in Enterococcus faecalis by Bioinformatics
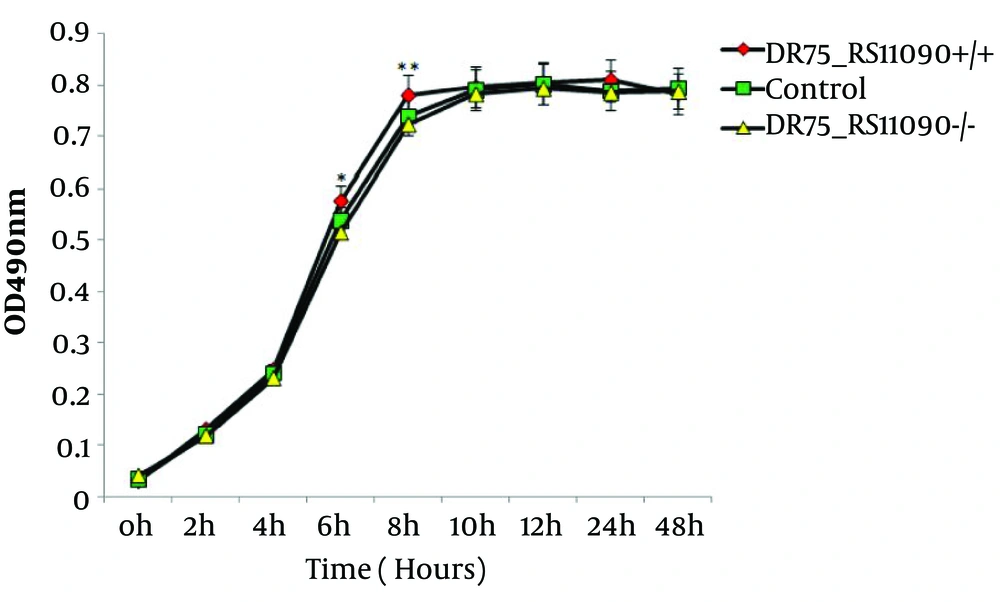
The genome was extracted from online NCBI database and analysis and comparison revealed that DR75-RS11090 gene (wp 002356022.1) had a conservative GGDEF domain (Figure 1). Next, DR75-RS11090 was overexpressed and knocked down in E. faecalis. The growth curve of E. faecalis was detected by MTT assay. DR75-RS11090 promoted E.faecalis proliferation. The test results are shown in Figure 2. The proliferative capacity of the three strains was improved with the increase of culture time. After 12 hours of culture, the number of bacteria reached the maximum. DR75-RS11090+/+ strains had the best proliferative capacity.
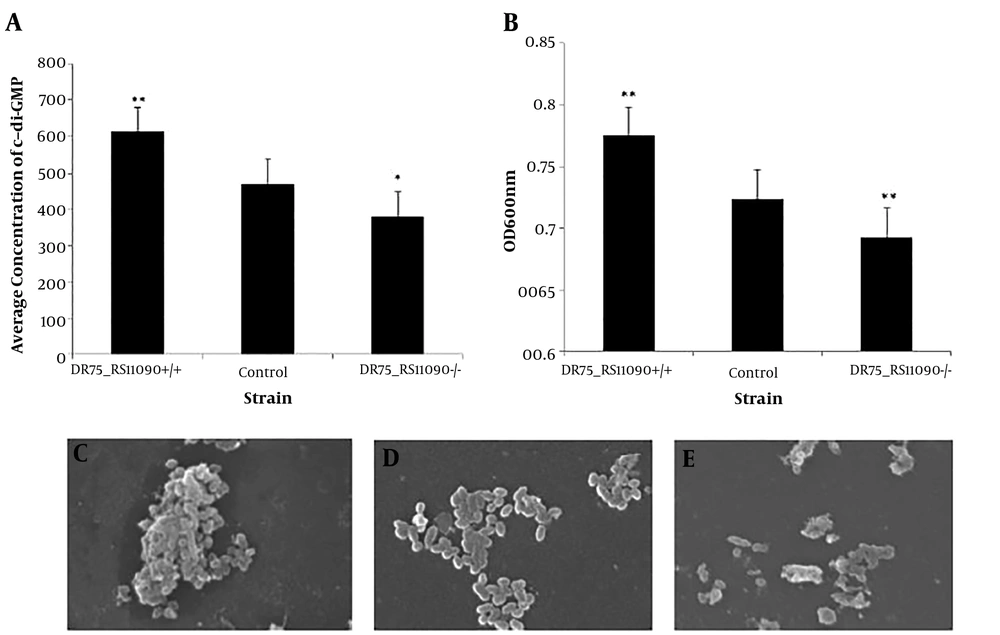
4.2. DR75_RS11090 Increased the Synthesis of c-di-GMP and Biofilm Formation in Enterococcus faecalis
The concentration of c-di-GMP was detected by High-performance liquid magneto-chromatography (HPL-MC). Overexpression of DR75-RS11090 in E. faecalis increased c-di-GMP synthesis (Figure 3A). The biofilm formation was represented by the light density values. The higher value of the light density represents the more living bacteria in the biofilm, and the stronger biofilm formation ability. DR75-RS11090+/+ strain had the largest optical density, suggesting that it had the strongest ability to form biofilm (Figure 3B). The morphology of E. faecalis was captured by the scanning electron microscope (SEM). As shown in Figure 3C, E. faecalis gathered into a colony, had clear morphology, and smooth membrane. The adhesion ability of the normal strain is general, and the individual morphology of the bacteria were clear, smooth, and without diamond angle (Figure 3D). DR75-RS11090-/- strains did not form biofilm, the bacteria were disrupted, and the individual bacteria were in poor state (Figure 3E). The results showed that DR75-RS11090 increased the synthesis of c-di-GMP and biofilm formation in E. faecalis.
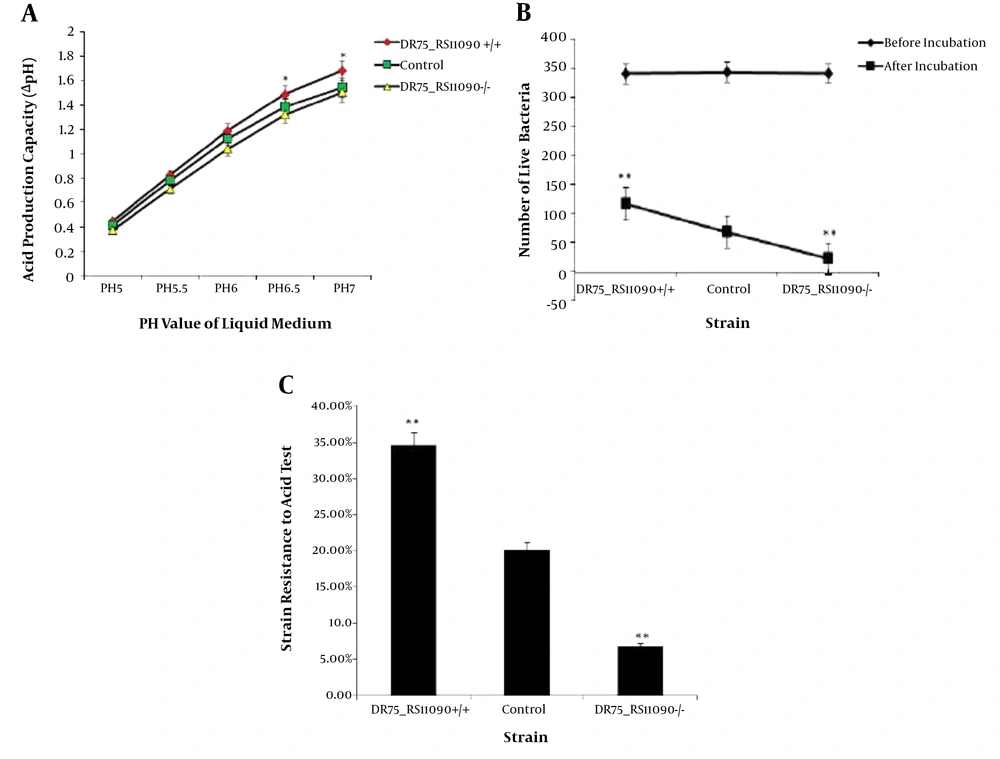
4.3. Detection of Acid-Producing and Acid Resistance Capacity of Enterococcus faecalis Strains
Acid-producing capacity was presented by ΔpH of the medium. The initial pH values of BHI medium ranged 5 to 7. The acid-producing ability of E. faecalis strains increased with ΔpH. DR75-RS11090+/+ strains had the largest ΔpH, while DR75-RS11090-/- had the smallest ΔpH (Figure 4A). It suggested that DR75-RS11090 promotes the acid-producing capacity. To detect the acid resistance capacity, the three groups of strains were cultured in an acidic medium (pH = 5) and incubated for two hours. The number of live bacteria in the three groups of strains decreased significantly (Figure 4B). DR75-RS11090+/+ strain had the most live bacteria, while DR75-RS11090-/- had the least (Figure 4C). It showed that DR75-RS11090 gene was important for E. faecalis to resist the acid.
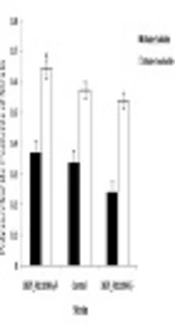
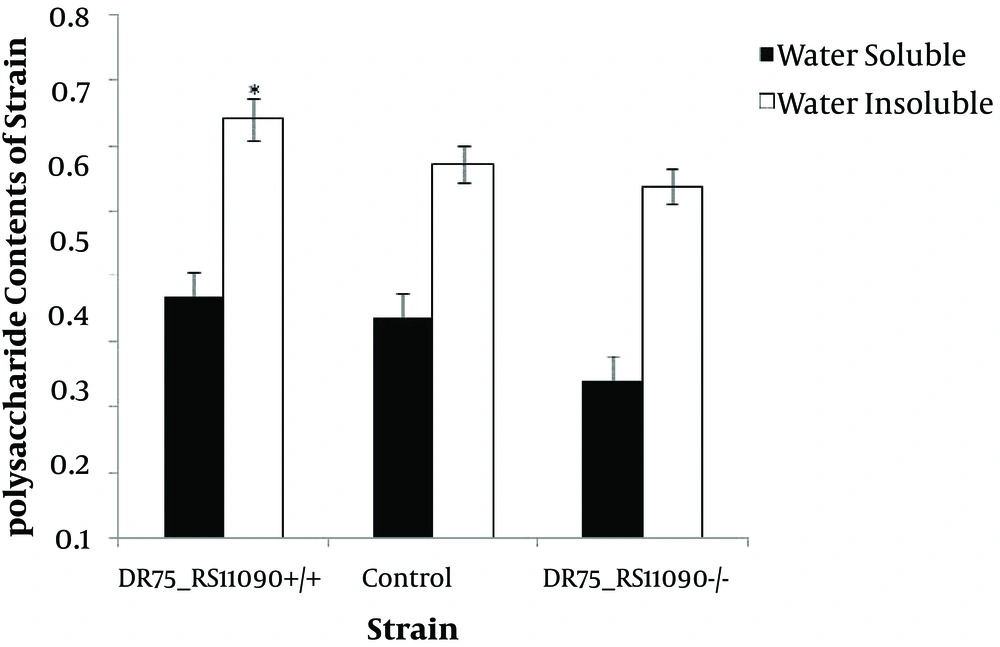
4.4. Determination of Water-Insoluble and -Soluble Polysaccharide Synthesis Ability
Water-soluble and -insoluble polysaccharide synthesis capacities of the three strains are shown in Figure 5. Enterococcus faecalis were better at producing water-insoluble polysaccharides than water-soluble ones. DR75-RS11090 increased the water-insoluble and -soluble polysaccharide synthesis ability.
5. Discussion
Many studies show that E. faecalis strains play an important role in intestinal microecological balance (10-13). Enterococcus faecalis regulates the balance of intestinal microecosystem (18). Imbalance of intestinal flora declines the immunity, resulting in various diseases. When intestinal microecosystem is disturbed, the number of bifidobacterium and lactobacillus decreases, while the number of Salmonella spp. and Escherichia coli increases (19). At this time, E. faecalis produces lactic acid to inhibit pathogenic bacteria growth. Enterococcus faecalis strains inhibit the adhesion of pathogenic bacteria by changing the composition of intestinal mucus and stimulating the body's immunity. Enterococcus faecalis strains are lactobacilli that can produce bacteriocins, organic acids, or volatile fatty acids to improve the intestinal environment (20, 21).
Bacterial biofilm can be regarded as the unstructured accumulation of bacterial cells surrounded by surface polysaccharide. The dehydrated biofilm is irregular when watching through an electron microscope (22). In the current study, a SEM was used to record the biofilm of the three groups. The morphology of the biofilm was clearly observed. Scanning electron microscope is widely used to observe biofilm morphology. Hickman and Harwood analyzed biofilm morphology, biomass, and cell distribution by a SEM and found that extracellular electron transfer promoted E. faecalis biofilm metabolism (14).
In the current study, colonies of DR75-RS11090+/+E. faecalis was found, which their biofilm was smooth and clear. It is proved that DR75-0RS11090 promotes the formation of biofilm. DR75-RS11090 contains GGDEF domain, which mediates the synthesis and activation of c-di-GMP. Subsequently, c-di-GMP promotes the formation of biofilm in E. faecalis (15). Romling et al. found that mutations in the GGDEF domain result in the loss of biofilm forming capacity (15). The c-di-GMP regulates various cellular processes including biofilm formation, motility, and virulenc (16). The c-di-GPM is the second messenger in bacteria. It stimulates the biosynthesis of adhesion and extracellular polysaccharide matrix in biofilms and controls bacterial motility. The c-di-GMP is produced by two GTPs (guanosine triphosphate) through guanosine acyclic cyclase. The activity of guanosine acid-cyclase is related to GGDEF domain (3).
The synthesis and degradation of c-di-GMP are mediated by the ornidazole (containing GGDEF domain) and phosphodiesterase (containing EAL or HD-GYP domain) (23). GGDEF, EAL, and HD-GYP domains were the first identified c-di-GMP modules using bioinformatics methods. High levels of intracellular c-di-GMP promote biofilm formation, while low levels lead to biofilm diffusion and return to plankton stages. Guanosine acyclozyme increases the level of c-di-GMP, and phosphodiesterase decreases its level. The current study showed that DR75-RS11090 promotes c-di-GMP synthesis and biofilm formation in E. faecalis. The current study result was consistent with that of the previous study on Salmonella spp. Simm et al. indicated that GGDEF proteins of Salmonella are regulated by c-di-GMP and affect the biofilm formation and cellulose production in Salmonella (24).
Enterococcus faecalis ferment carbohydrate from daily life diet and produce lactic acid. In mouth, E. faecalis strains cause tooth demineralization (25). Enterococcus faecalis produce acid to form an acidic environment. Other bacteria cannot grow in this acidic environment. However, E. faecalis strains have a strong acid resistance capacity. The c-di-GPM plays an indispensable role in the formation of E. faecalis biofilm. Low concentration of c-di-GMP inhibits biofilm formation and surface adhesion production, leaving bacteria in a swimming state. High concentration of c-di-GMP improves the expression of adhesive matrix components, generates multi-cell behavior, and promotes biofilm formation leaving the bacteria in a static state. Rao et al. found that YybT was a signaling protein containing GGDEF domain. They further demonstrated that YybT participated in acid resistance in Bacillus subtilis (26). In the current study, E. faecalis acid-producing ability was verified by pH value, and the number of living bacteria in acidic environment verified their acid resistant ability. DR75-RS11090+/+E. faecalis strain has the best acid-producing and acid resistant ability. It represents that DR75-RS11090 gene increases the acid producing and acid resistant ability in E. faecalis, which was consistent with the result of Rao’s research on B. subtilis.
There are two kinds of extracellular polysaccharides, water-soluble and -insoluble polysaccharides (27).Water-insoluble extracellular polysaccharides have strong viscosity to avoid being dissolved. Water-insoluble extracellular polysaccharides are the main virulence factors in E. faecalis. It was confirmed that DR75_RS11090 promoted the production of water-insoluble polysaccharides.
5.1. Conclusions
In summary, DR75-RS11090 gene was identified as an encoding GGDEF domain. And it was found that DR75-RS11090 promoted c-di-GMP synthesis, biofilm formation, and acid-producing and acid resistance capacities in E. faecalis.