1. Background
Non-typhoidal Salmonella (NTS) is a major foodborne pathogen associated with various gastrointestinal diseases both in developing and developed countries (1). Its clinical manifestation ranges from a self-limiting gastroenteritis characterized by vomiting, abdominal pain, diarrhea to serious invasive diseases such as bacteremia, extra-intestinal infections and meningitis (2-4). The occurrence of typhoid fever is more prevalent in developing countries, while NTS infections occur worldwide (5).
The NTS include Salmonella enterica subspecies enterica serotype enteritidis, S. typhimurium, S. newport, S. heidelberg, S. javiana, S. saintpaul, S. montevideo, S. infantis, S. muenchen, S. bareilly, S. braenderup and S. thompson (6). Globally, an estimated 93757000 cases of gastroenteritis and 155000 deaths due to NTS occur each year (1). Infants and young children are more susceptible to NTS infection compared to other age groups, making them a high-risk population (7). About 5% of the NTS infection in high-income countries occurs due to extra-intestinal and invasive NTS (iNTS). The iNTS found in the bloodstream of children is emerging with new pathogenic characteristics (8). Except for immunocompromised patients, the NTS infection does not necessitate antibiotic treatment, while, the iNTS infection requires an antibiotic treatment (9, 10). Similar to other enteropathogens, NTS exhibits an alarming level of increased resistance to various antibiotics especially to those used in developing countries (11, 12).
Resistance to first-line antibiotics including trimethoprim/sulphamethoxazole, chloramphenicol and ampicillin is high among the NTS. Thus, the treatment regimen mainly depends on fluoroquinolones or third generation cephalosporin’s. However, the decreased susceptibility to fluoroquinolones and extended-spectrum beta-lactamases (ESBLs) hampered NTS treatment. According to the US National Antimicrobial Resistance Monitoring System, about 5% of all NTS isolates were reported to be resistant to ceftriaxone (13). The rise of ciprofloxacin resistance has also been reported in both the US (27%) and China (5.5%) (14, 15).
The spread of antibiotic resistance makes it difficult to treat infants and children due to limited availability of therapeutic options, raising serious public health concerns (11, 16, 17). Antibiotic resistance especially due to extended-spectrum cephalosporins has been considered to be a major problem in various geographical regions including North/Western, Asia, Africa, US and Europe (18). In the last decade, the CTXM enzymes (blaCTX-M) which hydrolyse cefotaxime and ceftriaxone and the AmpC β-lactamase (blaCMY gene) which mediates resistance to other b-lactams, chloramphenicol, sulphonamides streptomycin, and tetracycline has gained much importance (18). In Europe and the USA, blaSHV, blaOXA, blaTEM, and blaCTX-Mare the frequently observed ESBLs among NTS (19). The spread of antibiotic resistance and the disease burden caused by NTS especially in children is on the rise worldwide, however, reports from China regarding the antibiotic resistance of NTS is scarce.
2. Objectives
The aim of this study was to determine the prevalence, antibiotic susceptibility of NTS and iNTS isolated from stool and blood samples of children aged 2 - 15 years, respectively.
3. Methods
A total of 537 stool samples and 405 blood samples were collected from 613 children with symptomatic gastroenteritis attending The Second Hospital of Lanzhou University, Lanzhou, China between January 2016 and May 2018. Children who had received prior antibiotic therapy within the last two weeks were excluded from the study. An informed consent from each child’s legal heir was obtained. Stool samples were collected using a sterile container, blood samples were collected in blood culture bottles (BD BACTECTM, USA). The collected stool and blood samples were immediately transported to the laboratory.
3.1. Isolation and Identification of Non-typhoidal Salmonella
The collected stool samples were cultured on MacConkey agar (Sigma-Aldrich, USA) and xylose lysine deoxycholate agar (Sigma-Aldrich, USA) and incubated at 37°C for 18 - 24 hours. Non-lactose fermenting colonies from MacConkey and pink colonies with a black center from xylose lysine deoxycholate agar plates showing typical colony morphology were isolated. Blood culture was done using an automated blood culture system (BD BACTECTM 9240, USA). The isolates were then identified through API® Rapid ID 32 E (bioMerieux, USA). The isolates were stored in brain heart infusion broth (Sigma-Aldrich, USA) supplemented with sterile glycerol (20%, v/v) at -80°C until used.
3.2. Minimum Inhibitory Concentration Assay
The minimum inhibitory concentration (MIC) for various antibiotics such as gentamicin, amikacin, ampicillin, cefotaxime, amoxicillin, ciprofloxacin, chloramphenicol, co-trimoxazole and nalidixic acid (Sigma-Aldrich, USA) against NTS was determined. The inoculum was prepared by adjusting the turbidity of the overnight culture grown on nutrient agar to that of 0.5 McFarland’s standard. The MIC assay (micro broth dilution method) was performed at a concentration of each antibiotic ranging from 0.03 to 128 µg/mL; using Muller-Hinton broth (MHB, Sigma-Aldrich, USA) as described in Clinical Laboratory Standard Institute (CLSI) guidelines (20). Briefly, 10 µL of culture was inoculated into MHB containing various concentrations and incubated at 37°C for 24 hours. After incubation, MIC was determined visually by the highest concentration showing the absence of growth.
3.3. DNA Extraction
DNA was extracted from the isolated cultures by the alkali lysis method (21). Briefly, a single colony of NTS was suspended in 100 µL of 50 mM NaOH and incubated at 95°C for 1 minute then immediately cooled to 4°C, neutralized with 16 µL of 1 M Tris-HCl (pH 8.0). Then the suspension was subjected to centrifugation for 2 minutes at 14000 rpm, the supernatant was collected and stored at -20°C until further use.
3.4. Resistance Gene Detection
A multiplex PCR to detect extended-spectrum β-lactamases (ESBL)-MBL resistant genes such as blaTEM, blaCTX-M-15, blaOXA, blaSHV and blaCMY-2was performed (22). Briefly, a 50 μL PCR reaction mix was prepared using a 2X ready PCR mix (Thermo Fisher Scientific, USA), 5 μL of template DNA and 5 pmoL of each primer as described in Table 1. PCR was performed using the thermocycler (Applied Biosystems, Verti Thermal Cycler, Thermo Fisher Scientific, USA), the PCR cycling conditions included: an initial denaturation at 94°C for 3 minutes followed by 30 cycles at 94°C for 30 seconds, appropriate primer annealing temperature for 30 seconds, extension at 72°C for 1 minute and final extension at 72°C for 7 minutes. After PCR, amplicons were resolved in 1.2% agarose gel electrophoresis.
| Gene | Primer Sequence (5’ - 3’) | Annealing Temperature (°C) | Amplicon Size (bp) |
|---|---|---|---|
| blaCMY-2 | F: AACACGGTGCAAATCAAACA | 58 | 332 |
| R: CCGATCCTAGCTCAAACAGC | |||
| blaCTX-M-15 | F: CCAGAATCAGCGGCGCACGA | 64 | 587 |
| R: GCGCTTTGCGATGTGCAGCA | |||
| blaSHV | F: AAGATCCACTATCGCCAGCAGG | 59 | 319 |
| R: ATTCAGTTCCGTTTCCCAGCGG | |||
| blaOXA | F: ATGAAAAACACAATACATATCAACTTCGC | 55 | 820 |
| R: GTGTGTTTAGAATGGTGATCGCATT | |||
| blaTEM | F: ATGAGTATTCAACATTTCCG | 55 | 859 |
| R: ACCAATGCTTAATCAGTGAG |
Primer Sequence for the Detection of Resistance Genes
3.5. Statistics
Continuous/categorical variables were represented as mean/ranges and numbers/percentages, respectively. Student t-test and ANOVA were performed using MINITAB (MINITAB, Version 13) statistical software. A P value of < 0.05 was considered as statistically significant.
4. Results
4.1. Patients and Isolates
Of the 613 patients, 374 (61%) were male and 239 (39%) were female. The mean age was 7.2 ± 0.5 years (range 1.2 - 13 years) (Table 2). Of the 537 stool samples and 405 blood samples, a total of 213 (39.7%) and 165 (40.7%) Salmonella sp. were isolated, respectively which was significantly higher than other bacterial species (P = 0.03). The majority of the isolates (172 vs. 134) were isolated from children within the age group of 1 - 5 years. Of the tested stool and blood samples, 54 (10.1%) and 38 (9.4%) isolates were identified as non-typhoidal Salmonella, respectively. The NTS isolated from stool samples were designated as NTS isolates and those which were isolated from blood samples were designated as iNTS isolates. Salmonella typhimurium was the predominant species identified in both NTS (38.9%) and iNTS (47.4%) groups, followed by S. enteritidis (29.6%) in the NTS and S. enterica in the iNTS (26.3%) groups (Table 3).
| Description | No. of Patientsa |
|---|---|
| Male | 374 (61) |
| Female | 239 (39) |
| Mean age, y | 7.2 ± 0.5 (1.2 - 13) |
| Acute gastroenteritis | 216 (35.2) |
| Chronic diarrhea | 112 (18.3) |
| Watery diarrhea | 73 (11.9) |
| Vomiting | 214 (34.9) |
| Anemia | 268 (43.7) |
| Fever | 256 (41.7) |
Baseline Patient Characteristics
| Species | All NTS (N = 92) | NTS group (N = 54) | iNTS group (N = 38) |
|---|---|---|---|
| Salmonella typhimurium | 39 (42.4) | 21 (38.9) | 18 (47.4) |
| S. enteritidis | 23 (25) | 16 (29.6) | 7 (18.4) |
| S. enterica | 20 (21.7) | 10 (22.2) | 10 (26.3) |
| S. infantis | 8 (8.7) | 5 (9.3) | 3 (7.9) |
| S. dublin | 2 (2.2) | 2 (3.7) | 0 |
Distribution of Non-Typhoidal Salmonella Speciesa
4.2. Detection of Antibiotic Resistance
Of the overall 92 (NTS, 54; iNTS, 38) isolates tested for antibiotic susceptibility by MIC, the majority of the isolates were resistant to nalidixic acid (50, 54.3%) followed by ciprofloxacin (44, 47.8%), cefotaxime (38, 41.3%), gentamicin (36, 39.1%), amoxicillin (32, 34.8%), ampicillin (28, 30.4%), co-trimoxazole (28, 30.4%), amikacin (24, 26.1%) and chloramphenicol (17, 18.5%) (Table 4).
Of the 54 NTS isolates, the majority of the isolates were resistant to nalidixic acid 35 (64.8%) followed by ciprofloxacin (25, 46.3%), cefotaxime (21, 38.9%), gentamicin (20, 37%), amoxicillin (19, 35.2%), co-trimoxazole (18, 33.3%), ampicillin (16, 29.6%), amikacin (15, 27.8%) and chloramphenicol (9, 16.7%) (Table 4). We found that antibiotic resistance by NTS towards nalidixic acid was significantly higher than other tested antibiotics (ANOVA, F = 0.23; P < 0.05).
Of the 38 iNTS isolates, the majority of the isolates were resistant to ciprofloxacin (19, 50%) followed by cefotaxime (17, 44.7%), gentamicin (16, 42.1%), nalidixic acid (15, 39.5%), amoxicillin (13, 34.2%), ampicillin (12, 31.6%), co-trimoxazole (10, 26.3%), amikacin (9, 23.7%) and chloramphenicol (8, 21.1%) (Table 4). There was no significant difference in the presence of antibiotic resistance among the isolates (ANOVA, F = 0.01; P > 0.05). In comparison, the majority of the NTS and iNTS isolates were found to be resistant to nalidixic acid (64.8%) and ciprofloxacin (44.7%), respectively.
| Antibiotics | All (N = 92)a | Non-Typhoidal Salmonella (N = 54) | Invasive Non-Typhoidal Salmonella (N = 38) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistant Isolatesa | MIC50 (µg/mL) | MIC90 (µg/mL) | MIC GM (µg/mL) | Range (µg/ml) | Resistant Isolatesa | MIC50 (µg/mL) | MIC90 (µg/mL) | MIC GM (µg/mL) | Range (µg/mL) | ||
| Gentamicin | 36 (39.1) | 20 (37) | 8 | 64 | 22.63 | 0.5 - 64 | 16 (42.1) | 8 | 32 | 16 | 0.5 - 64 |
| Amikacin | 24 (26.1) | 15 (27.8) | 16 | 128 | 42.25 | 0.5 - 128≤ | 9 (23.7) | 16 | 128 | 42.25 | 2 - 128≤ |
| Ampicillin | 28 (30.4) | 16 (29.6) | 4 | 32 | 11.31 | 0.25 - 64 | 12 (31.6) | 4 | 64 | 16 | 1 - 64 |
| Amoxicillin | 32 (34.8) | 19 (35.2) | 8 | 64 | 22.63 | 0.5 - 128 | 13 (34.2) | 8 | 32 | 16 | 0.5 - 32 |
| Cefotaxime | 38 (41.3) | 21 (38.9) | 2 | 64 | 11.31 | 0.5 - 64 | 17 (44.7) | 2 | 8 | 4 | 0.06 - 8 |
| Ciprofroxacin | 44 (47.8) | 25 (46.3) | 4 | 8 | 5.65 | 1 - 16 | 19 (50) | 4 | 32 | 11.31 | 0.25 - 32 |
| Chloromphenicol | 17 (18.5) | 9 (16.7) | 2 | 32 | 8 | 0.5 - 64 | 8 (21.1) | 2 | 32 | 8 | 0.5 - 128 |
| Co-trimoxazole | 28 (30.4) | 18 (33.3) | 4 | 128 | 22.63 | 2 - 128≤ | 10 (26.3) | 4 | ≥ 128b | NA | 0.06 - 128≤ |
| Nalidixic acid | 50 (54.3) | 35 (64.8) | 32 | 64 | 45.25 | 8 - 64 | 15 (39.5) | 64 | ≥ 128b | NA | 0.12 - 128≤ |
Distribution of Antibiotic Resistance Among Non-Typhoidal Salmonella Isolates
4.3. Detection of β-Lactamase Genes
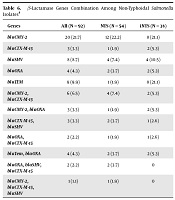
Of the β-lactamase genes tested in NTS and iNTS isolates, blaCMY-2was the predominant gene (33.3% vs. 31.6%) amplified by both the groups. The blaCTX-M-15and blaTEM were the next predominant genes amplified by NTS (20.4%) and iNTS (26.3%) isolates, respectively. There was no significant difference in the presence of resistant genes between the two groups (t-test, P > 0.05) (Table 5). Among these, blaCMY-2-blaCTX-M-15 was the common gene combination (7.4%) found in the NTS isolates while, blaCMY-2-blaCTX-M-15, blaCMY-2-blaOXA and blaTem-blaOXA were the predominant gene combination (5.3%) found in the iNTS isolates (Table 6).
| Genes | All (N = 92) | NTS (N = 54) | iNTS (N = 38) |
|---|---|---|---|
| blaCMY-2 | 30 (32.6) | 18 (33.3) | 12 (31.6) |
| blaCTX-M-15 | 17 (18.5) | 11 (20.4) | 6 (15.8) |
| blaSHV | 14 (15.2) | 9 (16.7) | 5 (13.2) |
| blaOXA | 13 (14.1) | 6 (11.1) | 7 (18.4) |
| blaTEM | 13 (14.1) | 3 (5.6) | 10 (26.3) |
Distribution of β-Lactamase Genes Among Non-Typhoidal Salmonella Isolatesa
| Genes | All (N = 92) | NTS (N = 54) | iNTS (N = 38) |
|---|---|---|---|
| blaCMY-2 | 20 (21.7) | 12 (22.2) | 8 (21.1) |
| blaCTX-M-15 | 3 (3.3) | 1 (1.9) | 2 (5.3) |
| blaSHV | 8 (8.7) | 4 (7.4) | 4 (10.5) |
| blaOXA | 4 (4.3) | 2 (3.7) | 2 (5.3) |
| blaTEM | 9 (9.9) | 1 (1.9) | 8 (21.1) |
| blaCMY-2, blaCTX-M-15 | 6 (6.5) | 4 (7.4) | 2 (5.3) |
| blaCMY-2, blaOXA | 3 (3.3) | 1 (1.9) | 2 (5.3) |
| blaCTX-M-15, blaSHV | 3 (3.3) | 2 (3.7) | 1 (2.6) |
| blaOXA, blaCTX-M-15 | 2 (2.2) | 1 (1.9) | 1 (2.6) |
| blaTem, blaOXA | 4 (4.3) | 2 (3.7) | 2 (5.3) |
| blaOXA, blaSHV, blaCTX-M-15 | 2 (2.2) | 2 (3.7) | 0 |
| blaCMY-2, blaCTX-M-15, blaSHV | 1 (1.1) | 1 (1.9) | 0 |
β-Lactamase Genes Combination Among Non-Typhoidal Salmonella Isolatesa
5. Discussion
Non-typhoidal Salmonella is associated with self-limiting gastroenteritis to serious invasive diseases (2-4). In developing countries, diarrheal illness is one of the major causes of morbidity and mortality in children (23). Salmonellosis is considered to be one of the most common infections associated with diarrheal illness posing a serious and significant public health problem especially in children (24). In our study, 18.3% of the children had chronic diarrhea and 11.9% had watery diarrhea. In our study, Salmonella sp. were isolated from 39.7% and 40.7% of the stool and blood samples, respectively. A study from Venezuela which included pediatric patients with acute gastroenteritis reported 15.2% of Salmonella sp. from stool samples which is lower than that reported in our study (25).
The incidence of NTS varies globally. In sub-Saharan Africa, which has a high prevalence of HIV and malarial infections, NTS was the common etiological agent for bacteremia (26). Africa suffers a high burden of NTS infection compared to Asia where the incidence was relatively low. In our study, the prevalence of NTS was 10.1% and iNTS was 9.4%. A study from Northwest China reported NTS prevalence at a rate of 3.75% from the diarrheal patients and 0.31% from the non-diarrheal patients, which was much lower than that reported in our study (27). In contrast, studies from the US (42%) and Shangai (17.2%) reported NTS as the leading cause of bacterial enteric illness, which was much higher than that reported in our study (15, 28). Similarly, a study from the West Indies reported a higher rate of NTS (21.1%) compared to our results (29).
A study from Bangladesh reported iNTS at a rate of 0.2%, which is much lower (9.4%) than that reported in our study, however, the study reported that of the 20 patients who had iNTS infection 5 patients died. The study also mentioned that clinical sepsis, severe acute malnutrition and pneumonia were independent risk factors for NTS bacteremia (30). Kariuki et al. reported that a larger proportion of the iNTS were isolated from children below 3 years which corroborates with our result where 52.9% of our iNTS isolates were from children less than 5 years old (16). Thus, the varying prevalence of NTS infection across various regions suggests that a region-specific approach is required to monitor the prevalence of NTS infections.
In our study, S. typhimurium was the predominant subtype identified in both NTS (38.9%) and iNTS (47.4%) groups. Similar to our results, studies from Kenya (59%) (16) and Iraq (54.5%) (23), reported that S. typhimurium was the predominant subtype of the NTS isolated, however with slightly higher rates. In our study, S. enteritidis (29.6%) was the second common subtype identified among the NTS isolated from stool samples. A study from Southwest China reported S. enteritidis as the most predominant subtype identified, however with a much lower rate (1.87%) than that reported in our study (27). Another study from Venezuela reported that S. enteritidis (48.7%) as the predominant serovar followed by S. typhimurium (37.8%) which is contrary to our result where S. typhimurium was the most common subtype followed by S. enteritidis (25). A study from India reported that S. enterica serovar senftenberg as the predominant isolate followed by S. enterica serovar typhimurium and S. enterica serovar enteritidis (22). Another study from West Africa reported S. enterica serovar enteritidis (80.6%), followed by S. enterica serovar typhimurium (8%) (31).
Although the predominance of NTS subtypes varies from one geographical region to other, S. typhimurium, S. enteritidis and S. enterica were the three major NTS subtypes commonly isolated from gastroenteritis patients. The SalmSurv, a WHO-supported foodborne disease surveillance network study reported that S. typhimurium and S. enteritidis cause approximately 80% of all the human cases, which corroborates with our results; where S. typhimurium and S. enteritidis were the major sub types identified in our study (32). An alarming increase in the antibiotic resistance by NTS was reported elsewhere (11, 12). In our study, 54.3% of our isolates were found to be resistant to nalidixic acid, which is lower than that reported from India (77%) (22), Southwest China (66.7%) (27), and higher than that reported from Congo (4.3%) (33), Iraq (45.5%) (23), Ethiopia (23.9%) (34), and Iran (31.5%) (17). Among the NTS isolated from stool samples, Nalidixic acid was predominant among our NTS, (64.8%) which was very much higher than that reported from Kenya (6%) (16). In contrast to our results, other studies from South India (8), and West Africa (31), reported that none of their NTS isolates were resistant to nalidixic acid.
In our study ciprofloxacin resistance (47.8%) was the second most common resistance reported, which was higher than that reported from Ethiopia (4.5%) (34), Congo (4.3%) (33), India (23.5%) (22), and lower than that reported from South India (97%) (8). Other studies from West Africa (31) and Malaysia (35) reported 100% susceptibility to ciprofloxacin. We reported that 41.3% of our isolates were resistant to cefotaxime, which was higher than that reported from India (32.5%) (22), Congo (2.1%) (33), Southwest China (11.9%) (27), and comparable to that reported from Iraq (42.4%) (23). Gentamicin resistance was reported to be 39.1% among our isolates, while studies from Ethiopia (7.5%) (34), Southwest China (11.9%) (27), reported lower and a study from India (40%) (22), reported a comparable rate of resistance to gentamicin. Other antibiotics for which our isolates were found to be resistant included amoxicillin (34.8%), ampicillin (30.4%), co-trimoxazole (30.4%), amikacin (26.1%), and chloramphenicol (18.5%), a study from Congo reported lower rates of resistance to the above-mentioned antibiotics (33). In comparison, a study from India reported a lower rate of resistance to amikacin (21%) and higher resistance to co-trimoxazole (33%) (22). Other studies reported that none of their isolates were resistant to chloramphenicol (23, 35).
Overall, the rate of antibiotic resistance is higher compared to several previous studies. We do not have specific data on the usage of antibiotics for the included patients in the current study. The increased resistance may be due to the fact that as our institute is a tertiary care hospital, there was every possible chance that the patients must be treated in a primary care setting and might be exposed to some antibiotics during the treatment at such centers. Drugs including β-lactams, aminoglycosides and fluoroquinolones have been used to treat various infections and the prolonged use of such drugs may lead to a high rate of antibiotic resistance.
The ESBL and the AmpC β-lactamase in Salmonella sp. have been reported in developing countries including India and Pakistan (36-38). However, data regarding the report of β-lactamase and the AmpC β-lactamase among Salmonella sp. is scarce in China. In our study, 32.6% of our isolates were positive for the blaCMY-2gene and significantly higher than other genes (t-test, P < 0.05). The blaCTX-M-15 was the next predominant gene found in our study. Similar to our study, a study from India reported that blaCMY-2(20.9%) was the predominant gene while blaCTX-M-15 was the next predominant gene amplified by their NTS, however, their rates were much lower compared to our results (22). In the current clinical settings, third-generation cephalosporins are the drug of choice for treating NTS infections, the presence of blaCTX-M-15, which spread among bacteria, especially the co-occurrence of plasmid-mediated ESBL and AmpC which can hydrolyse even carbapenems and pose a serious threat during the treatment of NTS infections. The study is limited by its retrospective and single-center design.
5.1. Conclusions
Overall, S. typhimurium was the predominant species identified and blaCMY-2 was the predominant gene amplified by our isolates. In general, invasive isolates exhibit less resistance, however, in our study the iNTS isolated from blood samples showed a similar resistance pattern to that of the NTS isolated from stool samples. Compared to other literature, the high prevalence and increased resistance especially among iNTS is a cause of concern and reiterates the need to test blood samples along with the stool samples for better management of gastroenteritis.