1. Background
Proteus species belong to the Enterobacteriaceae family (1). There are several species of Proteus including Proteus mirabilis, P. vulgaris, P. penneri, P. hauseri, P. myxofaciens (2, 3), P. alimentorum, P. cibarius, P. columbae, P. inconstans, P. morganii, P. terrae, and P. rettgeri (4-7). Proteus mirabilis is a Gram-negative, rod-shaped, facultative, anaerobic, non-capsulated, non-spore forming, and motile bacterium (8). It is mostly found in natural environments and responsible for the infection of the pulmonary system, burns, skin, eyes, ears, nose, and the urinary tract, as well as gastroenteritis (9, 10). Proteus mirabilis is the third most common cause of nosocomial infections accounting for 90% of all Proteus infections (11).
A major problem in wound infections is the ever-rising antimicrobial resistance in P. mirabilis (12-14). Proteus mirabilis may become resistant to β-lactams upon the acquisition of heterologous β-lactamase genes (15). A multidrug-resistant P. mirabilis clone with chromosomal AmpC-type beta-lactamase was reported in the literature (16). Thereby, P. mirabilis may be resistant to broad-spectrum β-lactams including penicillins and cephalosporins (17).
The virulence of Proteus spp. is dependent on the several virulent factors that are regulated by several genes encoded in operons (18). Proteus mirabilis utilizes the urease enzyme that plays an important role in the pathogenesis of kidney and bladder stone formation. Some genes such as ureA, ureB, ureC, ureD, ureE, ureF, ureG, and ureR on the ure operon are responsible for the production of the urease enzyme among which, ureC is a major contributor (19)
2. Objectives
The present study was designed to evaluate the different aspects of P. mirabilis and its antibiotic susceptibility pattern.
3. Methods
3.1. Collection of Wound Samples
The study was conducted from June 2017 to June 2018 at the Center for Advanced Studies in Vaccinology and Biotechnology (CASVAB), University of Balochistan, Quetta.
Overall, 480 wound samples (160 burn, 160 surgical, and 160 diabetic wounds) were collected from patients admitted to Bolan Medical Complex (BMC) Hospital. The samples were collected using pre-sterile pre-labeled cotton swabs and taken to the laboratory in cold chain condition. Data regarding the patients’ history, age, gender, and wound type were collected using predesigned checklists.
3.2. Sample Processing
The collected samples were streaked on differential (MacConkey agar, Oxoid) and selective (Proteeae isolation medium agar, Oxoid) media and incubated at 37ºC for 24 h. All isolates were triply cloned to get fresh colonies for Gram staining, biochemical tests (Indole, Methyl red, Voges-Proskauer, Simmon Citrate, Oxidase, and Catalase), and PCR.
3.3. Antimicrobial Susceptibility Test
Antimicrobial susceptibility was tested by the disc diffusion method according to the clinical and laboratory standards institute guidelines (CLSI guidelines, 2006). A suspension of bacterial cells (0.5 McFarland) was prepared and dispersed on the surface of Mueller-Hinton agar (Oxoid, UK) and incubated at 37ºC for 24 h. The isolates were assessed for sensitivity or resistance to particular antimicrobial agents such as penicillin G (10 μg), ampicillin (10 μg), amoxicillin (10 μg), cefuroxime (30 μg), ceftriaxone (30 μg), amikacin (30 μg), ceftazidime (30 μg), imipenem (10 μg), gentamicin (10 μg), ciprofloxacin (5 μg), and tetracycline (30 μg) based on inhibitory zones.
3.4. Polymerase Chain Reaction
The DNA was extracted from culture using a DNA purification kit (Hiper® Bacterial Genomic DNA Extraction Teaching Kit (India). After extraction, the DNA templates were stored at -20ºC for further use. The primer sequences including F: (CCG GAA CAG AAG TTG TCG CTG GA) and R: (GGG CTC TCC TAC CGA CTT GAT C) were designed to allow for the amplification of the 533-bp fragment of the ureC1 gene. For PCR amplification, a 25-μL volume reaction mixture was used containing 12 μL of master mix (2x AmpMaster™ Taq), 9 μL of grade water, 1 μL of each primer (forward, reverse), and 2 μL of template DNA. PCR cycling for reaction mixture included initial melting at 94ºC for 3 min, denaturing at 94ºC for 1 min, annealing at 63ºC for 30 s, extension at 72ºC for 1 min, and a final extension at 72ºC for 7 min. The final PCR product was run on the 1.5% agarose gel and observed under UV light.
4. Results
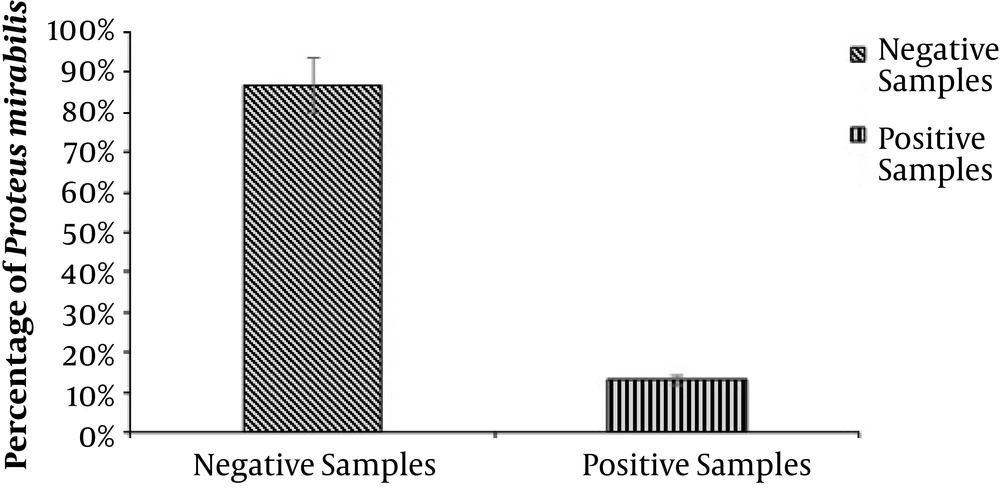
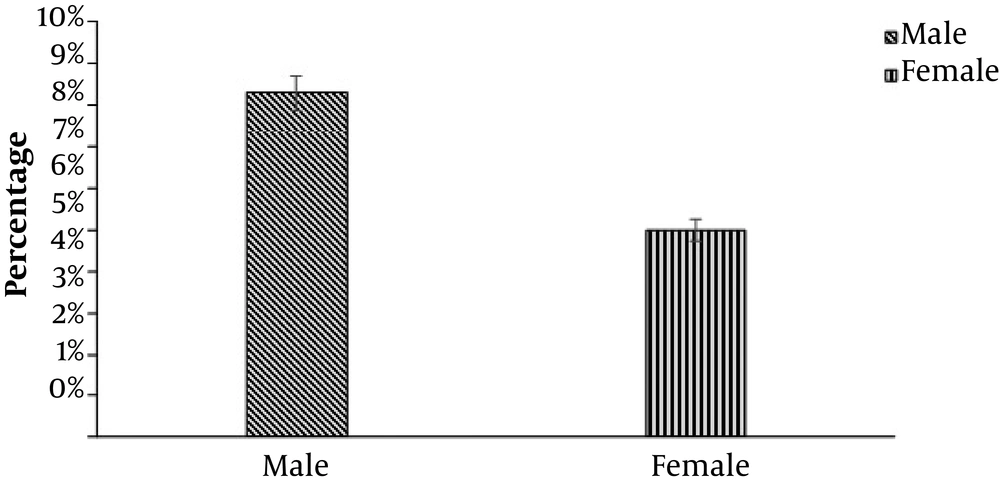
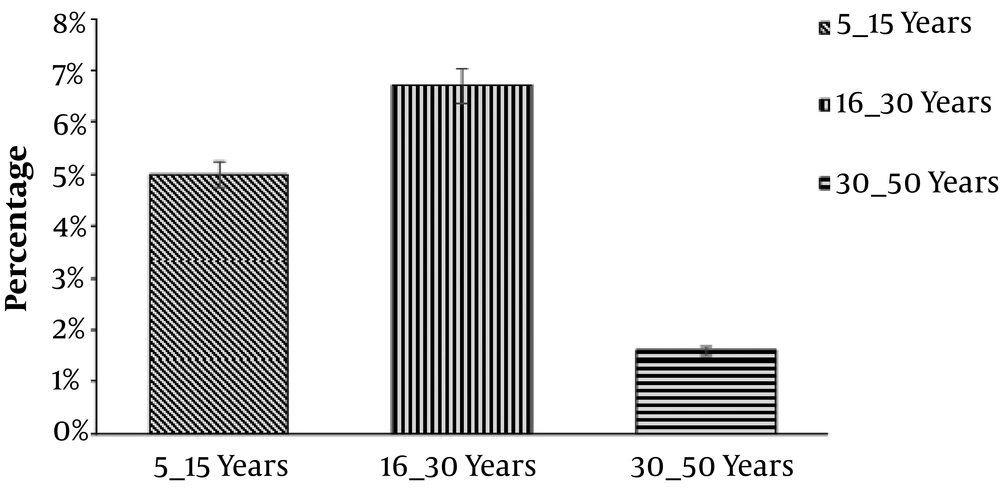
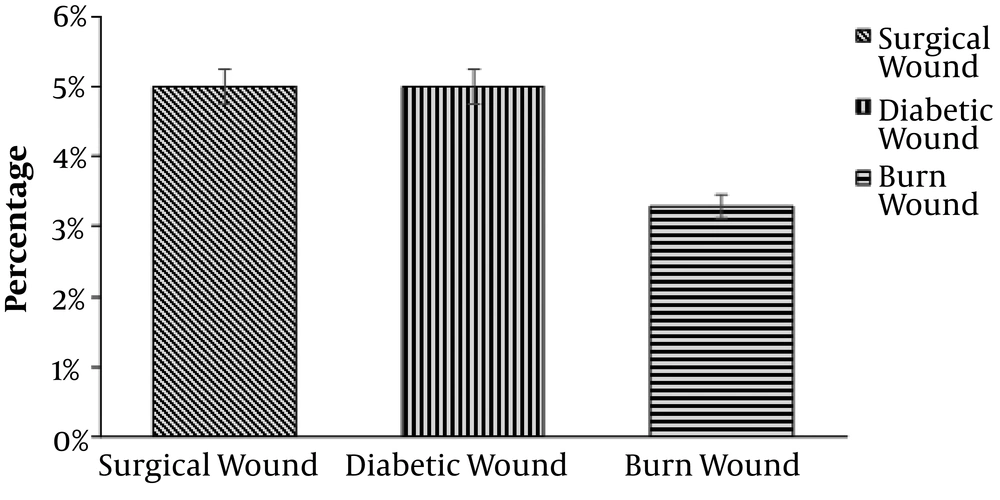
Overall, 480 wound samples were collected at Bolan Medical Complex Hospital, Quetta. There were 64 (13.3%) samples positive and 416 (86.6%) samples negative for P. mirabilis, as shown in Figure 1. Gender wise distribution showed that the wounds of male patients (n = 40; 8.3%) were more affected by P. mirabilis than the female patients’ wounds (n = 24; 5%), as shown in Figure 2. Wound infections caused by P. mirabilis affected all age groups including children, teenagers, and older people. According to our study, the rate of infection with P. mirabilis was the highest in 16 - 30-year-old patients (n = 32; 6.7%), followed by 5 - 15-year-old patients (n = 24; 5%) and 30 - 50-year-old patients (n = 8; 1.60%), as shown in Figure 3. The results also showed that diabetic (n = 24; 5%) and surgical (n = 24; 5%) wounds were more affected by P. mirabilis than burn wounds (n = 16; 3.30%), as shown in Figure 4. Proteus mirabilis was identified through differential and selective media, Gram-staining, and biochemical tests, as shown in Table 1.
| Proteus mirabilis | Values |
|---|---|
| Culture media | |
| MacConkey agar | Non-lactose fermenting, large, circular smooth colonies |
| Proteeae isolation medium (PIM) agar | Dark brown colonies with a zone of clearing in the medium |
| Gram staining | |
| Gram staining | - |
| Shape | Rods |
| Size | 1 - 3 µm in length |
| 0.4 - 0.8 µm in width | |
| Biochemical tests | |
| Motility | + |
| Indole | - |
| Citrate utilization | +/- |
| MR | + |
| VP | - |
| Urease | + |
| Catalase | + |
| Oxidase | - |
| Sugar fermentation tests | |
| Glucose | + |
| Sucrose | - |
| Sorbitol | - |
| Trehalose | + |
| Lactose | - |
| Mannitol | - |
| Dulcitol | - |
| Xylose | + |
| Inositol | - |
aPositive (+), Negative (-), Variable (+/-).
4.1. Antimicrobial Susceptibility Test
Proteus mirabilis isolates were sensitive to gentamicin (n = 50; 78%) and amikacin (n = 53; 82.8%) while resistant to penicillin G (n = 58; 90%), ampicillin (n = 56; 87.5%), amoxicillin (n = 60; 93.7%), cefuroxime (n = 61; 95.3%), ceftriaxone (n = 57; 89%), ceftazidime (n = 59; 92.1%), imipenem (n = 62; 96.8%), ciprofloxacin (n = 55; 85.9%), and tetracycline (n = 59; 92%), as shown in Table 2.
| Classes | Antibiotics | Abbreviation | Resistant, No. (%) | Sensitive, No. (%) |
|---|---|---|---|---|
| Penicillin | ||||
| Penicillin G | PEN | 58 (90) | 6 (9.3) | |
| Ampicillin | AP | 56 (87.5) | 8 (12.5) | |
| Amoxicillin | AMP | 60 (93.7) | 4 (6.2) | |
| Cephalosporin | ||||
| Cefuroxime | CXM | 61 (95.3) | 3 (4.6) | |
| Ceftriaxone | CTX | 57 (89) | 7 (10.9) | |
| Ceftazidime | CAZ | 59 (92.1) | 5 (7.8) | |
| Imipenem | IMP | 62 (96.8) | 2 (3.1) | |
| Aminoglycoside | ||||
| Gentamicin | GEM | 14 (21.8) | 50 (78) | |
| Amikacin | AMK | 11 (17) | 53(82.8) | |
| Quinolones | ||||
| Ciprofloxacin | CIP | 55 (85.9) | 9 (14) | |
| Tetracycline | ||||
| Tetracycline | TET | 59 (92) | 5 (7.8) |
4.2. Confirmation of P. mirabilis Through PCR
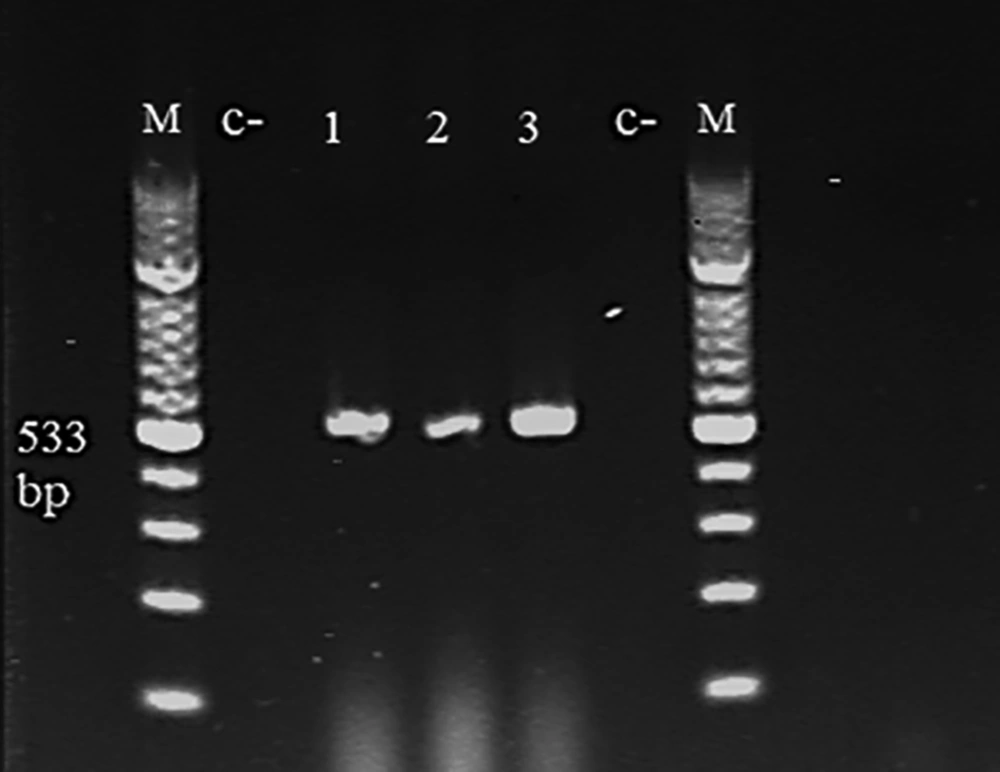
In the present study, the PCR-based identification of a specific gene was used to confirm P. mirabilis. All the isolates of P. mirabilis were used to produce a specific size of the 533-bp fragment of the ureC1 gene, as shown in Figure 5.
5. Discussion
Proteus mirabilis can cause both community and hospital-acquired infections, especially in immunocompromised patients. Based on the study by Brown (11), Proteus is the third main cause of hospital-acquired infections and a major contributor to the pathogenesis of wound infection. In the present study, 480 different wound samples were examined out of which, 13.3% were positive for P. mirabilis while 86.6% produced negative results. Among the positive samples, male patients’ wounds (8.3%) were more infected with P. mirabilis then female patients’ wounds (5%). Bahashwan and El Shafey (20) indicated in their study that males were more affected by P. mirabilis than females. This study showed that P. mirabilis affected all age groups, mostly 16 - 30 years-old (6.70%), followed by 5 - 10 years-old (5%) and 30 - 50 years-old (1.60%).
In this study, P. mirabilis was mostly found in surgical (5%) and diabetic (5%) wounds, followed by burn wounds (3%). Mohammed et al. (21) similarly reported in Nigeria that P. mirabilis was more frequently isolated from surgical wounds than burn and diabetic wounds. We identified P. mirabilis through differential media, Gram staining, and biochemical tests such as IMVIC, catalase, oxidase, sugar fermentation, citrate utilization, and urease and found results similar to the findings obtained by Cowan (22).
The control of wound infection has become more challenging due to the antibiotic resistance of infections. This study showed that P. mirabilis was sensitive to gentamicin and amikacin while resistant to penicillin G, ampicillin, amoxicillin, cefuroxime, ceftriaxone, ceftazidime, imipenem, ciprofloxacin, tetracycline. Mordi and Momoh (23) also found that all positive samples were resistant to tetracycline and erythromycin but sensitive to ciprofloxacin, ofloxacin, and gentamicin. In the present study, the PCR-based identification of a specific gene was used to detect P. mirabilis. All isolates of P. mirabilis were used to produce the specific size of the 533-bp fragment of the ureC1 gene; similar results were found by Abbas et al. (24).
Wound infection caused by P. mirabilis is a serious problem nowadays because bacteria have acquired resistance to many antibiotics. We should take those measures for the treatment of wound infections caused by P. mirabilis that can give us better results in the future. We also need to work on different antibiotic-resistance genes and study the whole genomic sequences of P. mirabilis using PCR.
5.1. Conclusions
The pathogenesis of P. mirabilis in wound infection and its antimicrobial sensitivity are major problems worldwide. The use of aminoglycosides such as gentamicin and amikacin is effective against P. mirabilis that helps prevent the spread of infection and reduce the cost of treatment. PCR is one of the sensitive, timesaving, specific, and cost-effective ways for the identification of the pathogenic genes of P. mirabilis.