1. Background
Helicobacter pylori as a Gram-negative facultative intracellular organism is able to move to the gastric mucosa due to its spiral form and leads to infection in gastric mucosa (1, 2). The prevalence of H. pylori infection is high and half of the world’s population is involved with this bacterium (3, 4). The colonization of this bacterium in the stomach is more likely to occur in people with chronic inflammation (5). Since most cases of H. pylori infection occur without any clinical symptoms, most people are unaware of this infection (3). However, the clinical manifestations, including gastroduodenal ulcer, gastritis, mucosa-associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma may be observed in most cases (6, 7).
Biopsy of H. pylori-infected individual has high level of immune cells infiltrate, including neutrophils, dendritic cells, monocytes, mast cells, and various types of lymphocytes and also pro-inflammatory cytokines, including interleukins (IL-1, IL-6 and IL-8), tumor necrosis factor-α (TNF-α), in comparison with uninfected people (8). In almost all individuals involved with H. pylori bacteria, the humoral and cellular immune responses occur simultaneously (9, 10). Microorganisms colonized in epithelial mucosa, such as H. pylori, are detected by pattern recognition receptors (PRRs) involved in the innate immunity. So, these receptors recognize the protected patterns found in many pathogens (11). NLRP3, is the most inflammasome complex that plays a critical role in inducing inflammation by recruiting adaptor proteins, and subsequently, triggers the caspase 1 protein, specific cysteine, and aspartate protease (12). The activation of caspase in the infected individuals is able to express certain inflammatory cytokines, including IL-1β and IL-18, in the dendritic cells; the presence of these two cytokines can lead to exacerbate inflammation (12-14).
NLRP3 gene is located on the short arm of chromosome 1 (1q44) and contains 9 exon codes (15). Some studies have shown that polymorphisms in PRRs genes may influence cytokine expression category in gastric mucosa of H. pylori-infected patients with gastritis (16). NLRP3 rs10754558 polymorphism has been associated with poor prognosis of coronary artery disease (CAD) in the Chinese population of Han. In addition, the G allele may be associated with an increase in the serum level of IL-1β (17). While other studies didn’t find any relationship between NLRP3 rs10754558 polymorphism and inflammatory diseases such as inflammatory bowel disease (IBD), Crohn disease, and gout (18, 19). The purpose of our study was to analyze the association of NLRP3 rs10754558 polymorphism with gastritis and peptic ulcer disease and to evaluate the effect of this polymorphism on the expression of IL-1β cytokine in H. pylori-infected patients.
2. Objectives
Since genetic variations such as polymorphisms may be involved in the expression of genes, this study was conducted to investigate the effect of NLRP3 rs10754558 polymorphism in pathogenesis of H. pylori infection.
3. Methods
3.1. Patients
To investigate the polymorphism of NLRP3 gene in patients with H. pylori-infection, 464 patients with dyspepsia participated in the study. Informed consent was obtained before biopsy sampling from the antral parts of the stomach. Five biopsy samples were obtained from the gastric antrum in each patient, which were then tested respectively for rapid urease, DNA extraction, RNA extraction, protein extraction, and histopathology study.
3.2. Detection of Helicobacter pylori
The histological examinations on samples from the antrum, the rapid urease test, and the 16S rRNA and glmM polymerase chain reaction (PCR) were used to detect the infections of H. pylori. Patients were considered H. pylori-infected only if all four tests were positive.
3.3. Histological Examination
First, the sections from biopsy samples were immersed in buffered formalin 10%. Then, Hematoxylin and Eosin staining method was used to stain the sections for detection of the gastritis. Moreover, a Giemsa stain was applied for H. pylori identification. An ulcer was defined as a limited mucosal impairment (with a diameter of > 5 mm and apparent depth) covered with exudates in the duodenum or stomach. The four-point Updated Sydney System (0: no; 1: mild; 2: moderate; and 3: severe changes) was utilized to classify and grade the severity of gastritis, ranging from normal to severe, according to the degree of mononuclear cell (MNC) and polymorphonuclear leukocytes (PMN) infiltration, and atrophy (20).
3.4. DNA Extraction and NLRP3 rs10754558 Genotyping
The samples from corpus were used to extract the genomic DNA with the aid of biospin tissue genomic DNA extraction kit (Bio Flux, Japan) acceding to the relevant protocol. A suspension was prepared from the extracted DNA in ultra-pure RNAse/DNAse-Free distilled water. The NLRP3 rs10754558 polymorphism was genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) as previously described by Li et al. (21).The thermal program of PCR process was a temperature of 95ºC for 5 minutes and then running 35 cycles at 95ºC for 30 seconds, at 58ºC for 30 seconds, at 72ºC for 30 seconds, and a final incubation at 72ºC for 5 minutes. In the next step, the digestion was performed on 6 μL-aliquot of the product using restriction enzyme MboI (Fermentas, Lithuania), followed by electrophoresis with 8% polyacrylamide gel to detect the NLRP3 rs10754558 alleles based on their size. Other laboratory personnel repeated the genotyping using randomly selected PCR samples in each genotype to verify genotyping results, which showed no difference.
PCR-RFLP approach was employed to detect the single nucleotide polymorphisms (SNP) of NLRP3. Single nucleotide polymorphisms of NLRP3 was detected using the PCR-RFLP procedure. PCR reaction was run with the set-up program: at 95ºC for 5 minutes, and then 35 cycles at 95ºC for 30 seconds, at 58ºC for 30 seconds, at 72ºC for 30 seconds, and a final incubation at 72ºC for 5 minutes. The primer for the PCR-RFLP was designed using the genomic sequences based on the Gen Bank (http://www.ncbi.nlm.nih.gov). The primer used for detection of SNP is shown in Table 1. To evaluate the accuracy of obtained results, several samples were selected and re-genotyped randomly.
| SNP | Primers Sequence | Annealing Temperature | Enzyme | Restriction Enzyme Digest Fragment Size, bp |
|---|---|---|---|---|
| rs10754558 | F: 5-CAGGACAATGACAGCATCGGGTGTTGAT-3 | 58.5ºC | MboI | GG: 261; CG: 261; 236; CC: 236 |
| R: 5-GCTGCCATAAAATTTCAACATAA-3 |
aPositive strand of nucleotide sequence was used to design primers for rs10754558. For the forward primer, a single nucleotide artificial mismatch was introduced within the three bases closest to the 3’end (GTT → GAT) to produce an MboI site.
3.5. Quantitative Analysis of IL-1β mRNA in the Gastric Mucosa Using Real-Time PCR
In this study, 75 patients were selected to evaluate the level of IL-1β in the gastric mucosa, which included 25 patients with GG genotype, 25 patients with GC genotype, and 25 patients with CC genotype of NLRP3 rs10754558 polymorphism. We took the same number of biopsies from patients with the ancestral allele (also called as wild allele), which were fitted to the control group by sex, age, and pathology. The total RNA extraction was carried out from the biopsy samples with the aid of a TRIzol® Plus RNA Purification Kit based on corresponding protocol. Following the treatment with DNase, 5 μg of total RNA was left with 2 μL of Super Script II reverse transcriptase (Revert Aid First cDNA synthesis (#K1632, Fermentase, Lithuania)). To normalize the results of cytokine mRNA expression, the house keeping gene of β-actin was used. The mRNA levels of cytokine and β-actin was quantified by the RotorGene Q cycler (Corbett Life Sciences; Doncaster, Australia) designed for IL-1β, and β-actin expression, shown in Table 2.
| Gene | Primers | Probes |
|---|---|---|
| β-actin | F: 5-AGCCTCGCCTTTGCCGA-3 | FAM-CCGCCGCCCGTCCACACCCGCC-TAMRA |
| R: 5-CTGGTGCCTGGGGCG-3 | ||
| IL-1β | F: 5-ACAGATGAAGTGCTCCTTCCA-3 | FAM-CTCTGCCCTCTGGATGGCGG-TAMRA |
| R: 5-GTCGGAGATTCGTAGCTGGAT-3 |
Reaction mixtures (25 μL) for PCR contained 1mg of synthesized cDNA solution with Taqman probe master mix (Qiagen, Germany). PCR was run at 95ºC for 10 minutes, and then 45 cycles of 95ºC for 15 seconds, and 60ºC for 60seconds, based on the manufacturer’s protocol. All experiments were repeated twice and all cytokine experiments were examined by analyzing all RNA samples in the same test. Relative quantification of cytokine to β-actin ratio (cytokine mRNA/β-actinm RNA) was calculated in accordance with the 2-ΔΔCт method (22).
3.6. SDS-PAGE and Western Blotting
Proteins were extracted using RIPA buffer. The samples after applying on the gel electrophoresis were transferred onto methanol-treated polyvinylidine difluoride membranes with the aid of Trans-blot cell transfer system (Bio-Rad, USA). Then, the immunolabeling procedure was carried out on the resulting membranes using rabbit anti-human polyclonal antibodies against β-actin (1:5000; Abcam, England). Also, secondary antibody used in this test was goat anti-rabbit IgG antibody coupled with HRP (1:1000; Abcam, England). The probe of membranes was done in line with the pierce ECL western blotting substrate instruction (Thermo Scientific; Scoresby, Australia).
3.7. Statistical Analysis
The statistical analysis of the polymorphism frequency was performed by Fisher’s exact test among the different groups. The odds ratio (OR) was calculated to evaluate the association of the polymorphisms and the diseases and the results were expressed as OR with 95% confidence interval (CI). The expression of cytokine was reported as mean and standard deviation. The independent t-test was used to analyze the differences between groups with and without polymorphism. In all tests, P value of 0.05 was statistically considered as significance level.
4. Results
4.1. Demographic Characterization of Studied Population
The general profiles of the studied population are shown in Table 3. Helicobacter pylori-infection was detected in 64.7% of the subjects. The statistical analysis of the groups provided the following results: the Helicobacter pylori-positive group with gastritis (n = 237 with 110 males and 127 females and the mean age of 50.18 ± 15.02 years); H. pylori-positive group with peptic ulcer diseases (n = 63 with 35 males and 28 females and the mean age of 50.16 ± 15.3 years); H. pylori-negative group with non-ulcer dysplasia (n = 164 with 75 males and 89 females and the mean age of 50.96 ± 19.77 years).
| Characteristics | Patients | Control Subjects | |
|---|---|---|---|
| Gastritis | Peptic Ulcer Diseases | ||
| Male/female, No. | 110/127 | 35/28 | 75/89 |
| Age, y (mean ± SD) | 50.18 ± 15.02 | 50.16 ± 15.3 | 50.96 ± 19.77 |
| Total, No. (%) | 237 (51) | 63 (13.5) | 164 (35.3) |
4.2. Polyacrylamide Gel Electrophoresis of PCR-RFLP to Determine the NLRP3 rs10754558 Genotyping
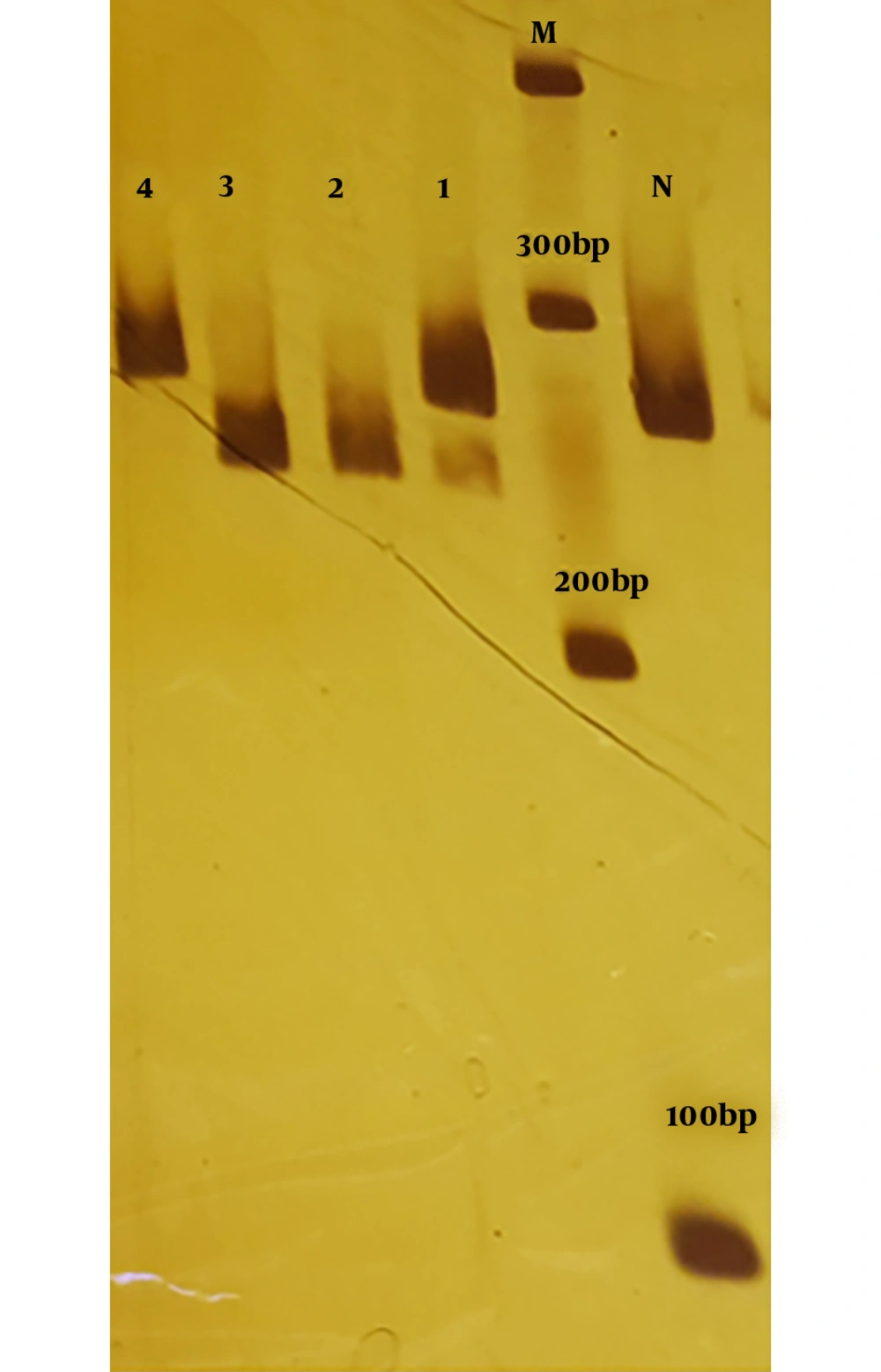
PCR products was digested by MboI enzyme to distinguish the NLRP3 rs10754558. The product length of PCR reaction for rs10754558 was 261 bp. For homozygous wild-type (GG), the enzyme does not work, and there will be a single band with 261 bp. For homozygous mutant genotype (CC), the enzyme cuts the two alleles, and the PCR product will appear as a single band of 236 bp, whereas the heterozygous genotype (GC) will appear as two bands of 261 and 236 bp (Figure 1).
4.3. Evaluation of NLRP3 rs10754558 Genotyping and Helicobacter pylori Infection Susceptibility
The frequency of three genotypes of NLRP3rs10754558 was assessed in both patient and control groups. The results are shown in Table 4. No significant difference was observed in the genotype of the polymorphism NLRP3rs10754558 between infected individuals and control subjects. Among the three genotypes of polymorphism, the frequency of GC was higher than the other two in both patients and control groups.
| NLRP3 (rs10754558) Genotype | Helicobacter pylori Infected, No. (%) | H. pylori Uninfected, No. (%) | P Value | OR (95% CI) |
|---|---|---|---|---|
| CC | 71 (23.7) | 44 (26.8) | 0.451 | 0.846 (0.547 - 1.308) |
| CG | 148 (49.3) | 82 (50) | 0.891 | 0.974 (0.665 - 1.425) |
| GG | 81 (27) | 38 (23.2) | 0.367 | 1.226 (0.787 - 1.911) |
4.4. Evaluation NLRP3 rs10754558 Polymorphism and Susceptibility to Gastritis and Peptic Ulcer Diseases in Helicobacter pylori-Infected Patients
Frequency of different genotypes of NLRP3rs10754558 polymorphism based on H. pylori infection and the type of disease caused by it are presented in Table 5. Statistical data indicated no significant association of the polymorphism frequency and the disease types in infected subjects with H. pylori.
| NLRP3 (rs10754558) Genotype | Infected Patients with Gastritis, No. (%) | Infected Patients with PUD, No. (%) | P Value | OR (95% CI) |
|---|---|---|---|---|
| CC | 56 (23.6) | 15 (23.8) | 0.976 | 0.990 (0.515 - 1.902) |
| CG | 115 (48.5) | 33 (52.4) | 0.584 | 0.857 (0.491 - 1.494) |
| GG | 66 (27.8) | 15 (23.8) | 0.521 | 1.235 (0.648 - 2.355) |
4.5. Evaluation of NLRP3 rs10754558 Polymorphism and Cellular Infiltration in Helicobacter pylori-Infected Patients
In current study was observed the relationship between NLRP3 (rs10754558) polymorphism and the rate of migration of inflammatory cells, which points out acute and chronic inflammation in the infected individuals with H. pylori (Table 6). The degree of acute inflammation in infected individuals with H. pylori with GG genotype was higher than those infected with GC and CC genotypes (P = 0.016), however; no significant correlation was found between NLRP3 rs10754558 polymorphism and chronic inflammatory severity (P = 0.979).
| NLRP3 (rs10754558) Genotype | Chronic Inflammation | Acute Inflammation |
|---|---|---|
| CC | 2.04 ± 0.155 | 0.74 ± 0.174 |
| CG | 2.04 ± 0.107 | 1.15 ± 0.107 |
| GG | 2.07 ± 0.103 | 1.37 ± 0.152 |
| P value | 0.979 | 0.016 |
aValues are expressed as mean ± SD.
bThe rate of inflammation is in the range (0-3).
4.6. Mucosal IL-1β mRNA Levels in Gastric Mucosa
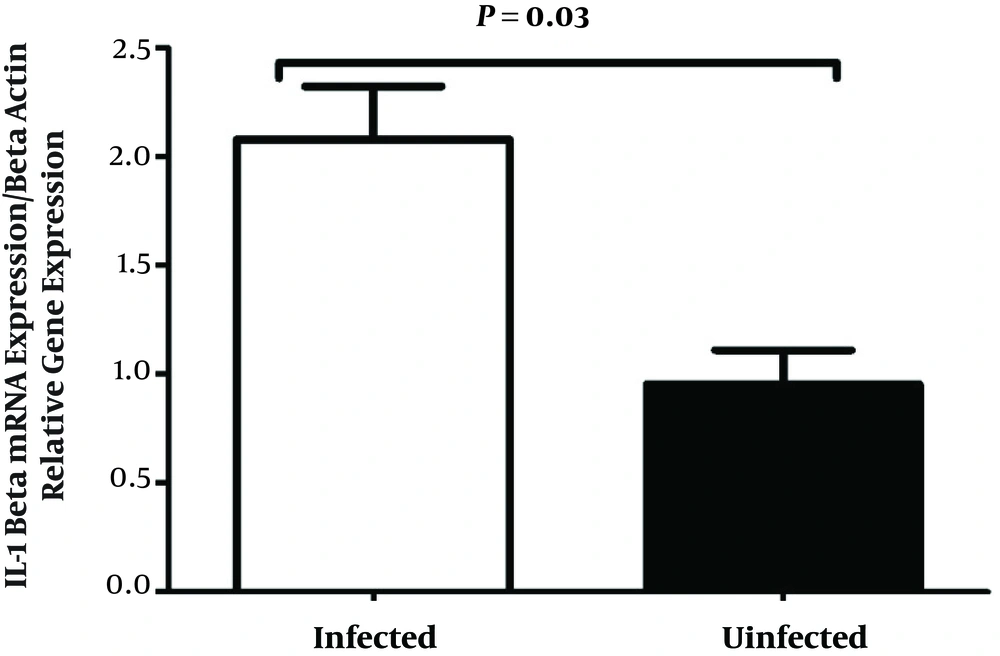
IL-1β mRNA levels was measured in two groups (Figure 2). IL-1β mRNA was elevated in infected subjects as compared to uninfected controls. In addition, the results showed that the expression of IL-1β in patients was 2.18-folded higher than that in the control subjects.
4.7. The Effect of NLRP3 rs10754558 Polymorphism on the Expression of IL-1β in Helicobacter pylori-Infected Patients with Gastritis and Peptic Ulcer Diseases
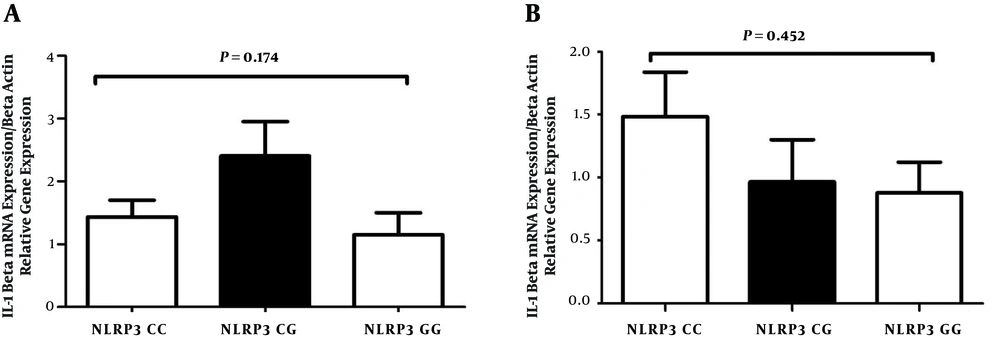
The function of NLRP3 genes might be influenced by the polymorphisms, so the hypothesis of the present study was that the polymorphisms might change the IL-1β expression in the gastric mucosa. Hence, the study evaluated whether the variants of NLRP3 allele’s modified the expression of IL-1β in infected patients with gastritis and peptic ulcer diseases. Our results in infected patients with gastritis and peptic ulcer diseases demonstrated no significant expression of IL-1β between infected patients with SNPs in NLRP3 and the wild-type allele (Figure 3).
The expression of IL-1β between infected patients with SNPs in NLRP3 and infected patients with the wild-type allele. IL-1β production levels are not significantly different in various genotypes of polymorphism in infected patients with gastritis (A) and peptic ulcer disease (B). The mRNA expression of IL-1β was evaluated using real-time PCR.
4.8. The Effect of NLRP3 rs10754558 Polymorphism on IL-1β Protein in Infected Patients with Gastritis
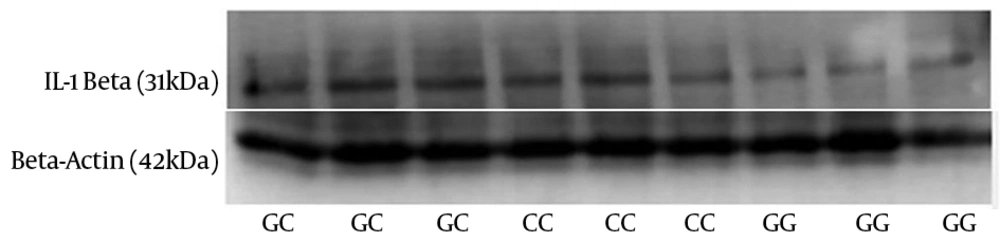
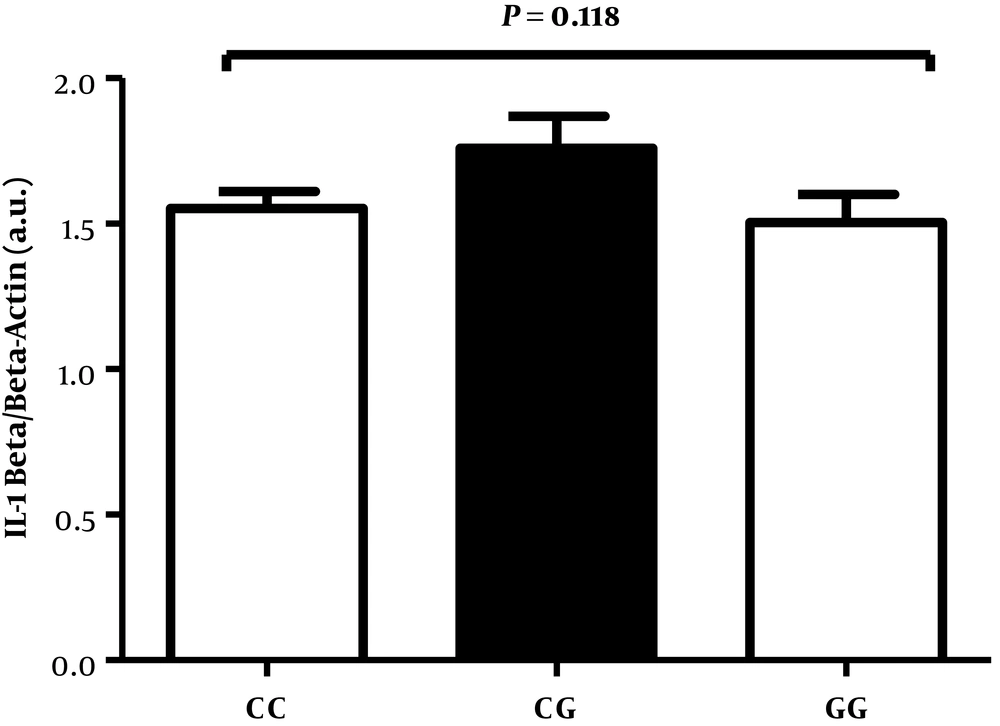
Western blotting was performed to detect the level of IL-1β protein in 75 infected patients with three different genotypes of polymorphism (25 subjects from each genotype). No significant differences were obtained between these 3 groups in infected patients with gastritis (Figures 4 and 5).
Expression of IL-1β by Western blot analysis in freshly obtained human gastric biopsies of H. pylori-infected patients with gastritis (25 subjects from each genotype). β-actin was used as loading control. Values are expressed as arbitrary units (a.u.). The horizontal bar represents the mean level of respective NLRP3genotype.
5. Discussion
Helicobacter pylori as an infectious agent in the gastric mucosa cause the infiltration of neutrophils, lymphocytes, monocytes and plasma cells. The results of previous studies have shown that the activation and the migration of inflammatory cells toward the gastric mucosa depend on the production of pro-inflammatory cytokines (7, 22, 23). On this note, molecular structures of inflammasomes, that play a prominent role in identifying pathogens and inciting stronger immune responses, are formed (24). According to reports, the polymorphisms in the NLRP3 gene could increase the susceptibility to some inflammatory diseases such as ulcerative colitis, juvenile idiopathic arthritis, inflammatory bowel disease, neonatal onset multisystem inflammatory disease, and type 2 diabetes (18, 25-29). Interestingly, in the present study, there were conflicting results with the aforementioned findings, such that there was no association between NLRP3 rs10754558 and H. pylori infections or diseases related to this microorganism.
Kim et al. (30) reported that H. pylori could induce the secretion of IL-1β in the innate immune cells, which symbolizes the NLRP3 inflammasome activity and needs the Cag Pathogenicity Island. The study of Castano-Rodriguez et al. (31) have shown the correlation between host genetic factors in gastric diseases caused by H. pylori infection, which revealed that the polymorphisms in genes involved in the nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway are related with infection caused by H. pylori among the ethnic Chinese people. They also showed the association of H. pylori strains with gastric cancer and gastritis, as well as found different levels of the expression of cytokines and chemokines. Helicobacter pylori, which causes the gastritis and peptic ulcer, is detected by PRRs, so the role of molecule involved in the NOD-like receptor (NLR) signaling pathway, NLRP3, was investigated in these diseases.
Zhang et al. (18) revealed that rs10754558 of NLRP3 is a crucial factor that can affect the development of ulcerative colitis in a Chinese population. Deng et al. (19) also reported the association of GG genotype of rs3806268 in NLRP3 gene with the elevated risk of primary gout in Chinese people. Zheng et al. (32) reported the overexpression of NLRP3 in aorta of patients suffering from coronary atherosclerosis and the correlation of aortic NLRP3 expression with the disease severity. Furthermore, Zhou et al. (17) showed that the G allele of NLRP3rs10754558 is related with the onset and poor prognosis of coronary artery disease (CAD) in Chinese Han population. They have also shown that the G allele susceptibility to CAD is associated with an increase in serum level of IL-1β in CAD. Current evidence suggests that polymorphisms involved in NLRP3 inflammasome may play a role in the prognosis or progression of some diseases.
Therefore, it was hypothesized that functional SNPs in NALP3 inflammasome may be involved in the severity of the type of diseases caused by H. pylori, by increasing or reducing cytokines secretion. At first, the expression of IL-1β and IL-18 genes between uninfected and H. pylori infected individuals were compared. It was found that these cytokines were incremented in the patients infected with H. pylori in comparison with healthy controls, consistent with studies of Hitzler et al. (13). They have shown the activation of caspase-1 by H. pylori and thus IL-1β/IL-18 secretion in dendritic cells (DCs) in the in vitro and also in the in vivo conditions. Two studies recently conducted on the H. pylori-infected human gastric cell lines indicated an increased expression of caspase-1, IL-1β, and IL-18 (33, 34).
Furthermore, no association was found between H. pylori infection and NLRP3-rs10754558, as well as between SNP and the type of diseases caused by H. pylori infection such as gastritis and duodenal ulcer. This was in agreement with other previous findings, which did not show any association between this polymorphism and diseases such as inflammatory bowel disease (IBD), Crohn disease, and gout (18, 19). While some other studies have shown the association between this polymorphism and diseases such as gastric cancer and ulcerative colitis (18, 31). Our findings show that NLRP3 rs10754558 polymorphism may be involved in the severity of some diseases caused by H. pylori infection.
5.1. Conclusions
Our findings show that rs10754558 polymorphism might not participate in regulating inflammation and immune responses in patients with H. pylori by influencing the expression of components of the NLRP3 inflammasome.