1. Background
Candida albicans is a predominant fungal opportunistic pathogen responsible for both mucosal and systemic infections, and is especially prevalent in immunodeficient hosts (1-4). Infections caused by C. albicans are mainly treated with azoles, which include both imidazoles (miconazole, clotrimazole, and ketoconazole) and triazoles (fluconazole and itraconazole). However, the long-term, repeated usage of azoles has resulted in the emergence of resistant isolates (5, 6). Drug resistance is a serious complication that can be challenging to clinicians, posing a major hurdle in the success of antifungal therapy (7). Previous studies have shown that decreased susceptibility to azoles is mainly associated with alterations in the ERG11 gene and/or constitutive upregulation of multidrug efflux pumps (8, 9). Azoles act as non‐competitive ERG11 inhibitors through coordination with the iron atom of the heme group located in the active site of 14‐α‐sterol demethylase.
Numerous publications have shown that non-synonymous point mutations in ERG11 may lead to conformational changes, resulting in the decreased affinity of azoles for the 14‐α‐sterol demethylase enzyme (9-11). In addition, elevated gene expression levels of efflux transporters can reduce the intracellular accumulation of certain drugs, and is often the main mechanism of azole resistance in clinical Candida (12). There are two families of membrane-associated efflux pumps, including ATP-binding cassette (ABC) superfamily (12) and major facilitator superfamily (MFS) (13), which have been found to be upregulated in resistant isolates. Two major ABC transporter genes, CDR1 (14) and CDR2 (Candida Drug Resistance1 and 2), are well-documented in clinical drug resistance. Both genes have shown to be upregulated in azoles-resistant C. albicans isolates and the genetic deletion of both genes in C. albicans results in hypersensitivity to azoles (15-19).
The expression levels of MDR1 (Multidrug resistance1) and FLU1 (Fluconazole resistance1), which are the members of the MFS, have also shown to be specifically upregulated in azole-resistant C. albicans strains (20, 21). In addition, several other factors can contribute to C. albicans azole resistance, including processes involved in biofilm formation and mutations in other enzymes in the ergosterol pathway (22). More recently, a novel gene (resistant to 7-aminocholesterol, RTA2), which has also shown to contribute to the development of azole resistance, has been identified in a mutant strain lacking the CDR1, CDR2, and MDR1 genes (23). Taken together, it is apparent that drug resistance is a multifactorial phenomenon. Due to the increased number of resistant strains, it is increasingly important to investigate the underlying mechanisms of azole resistance for the development of new antifungal treatments for C. albicans.
2. Objectives
In our previous study, we sequenced several fluconazole-resistant clinical isolates and found that 63.27% of the isolates did not contain mutated ERG11 (24). Our findings strongly suggested that other resistance mechanisms exist that require further research. In this study, we first analyzed the susceptibility of these isolates to other antifungal agents, and then investigated the relationship between fluconazole resistance, alternations in the ERG11 gene, and the expression levels of several membrane-associated efflux pumps (CDR1, CDR2, MDR1, FLU1) and RTA2 genes to decipher the potential molecular mechanisms leading to fluconazole-resistance in these clinical C. albicans strains.
3. Methods
3.1. Strains
We selected 40 fluconazole-resistant C. albicans isolates from our previous study using standard drug susceptibility analysis assays, defined herein as the fluconazole-resistant group. Additionally, 40 C. albicans isolates, which were susceptible to all drugs evaluated in our study, were defined as the susceptible group. All the C. albicans isolates were obtained from patients undergoing treatment for respiratory, genital, bloodstream, urinary, and digestive tract infections from 2005 to 2008 at the First Affiliated Hospital of Nanchang University in China. For drug susceptibility analysis, we used C. krusei ATCC 6258 and C. parapsilosis ATCC 22019 as quality control reference strains. Fluconazole-sensitive C. albicans ATCC 90028 was included as a control for the analysis of gene expression.
3.2. Antifungal Agents
Ketoconazole, itraconazole, 5-flucytosine, amphotericin B, clotrimazole, and nystatin were obtained from Sigma-Aldrich (USA). The measurable concentrations of ketoconazole, itraconazole, amphotericin B, clotrimazole, and nystatin ranged from 0.0313 to 16 μg/mL and dilutions of 5-flucytosine used herein ranged from 0.125 to 64 μg/mL.
3.3. Drug Susceptibility Analysis
The antifungal agent susceptibility analysis was performed according to the CLSI Broth Microdilution Susceptibility Method (M 27-A3 Document) for antifungal susceptibility of yeast. Briefly, stock inoculum suspensions of C. albicans were prepared and diluted with RPMI 1640 broth to obtain two inoculums (1 × 103 to 5 × 103 CFU/mL). Next, we plated 100 μL of each suspension into wells already containing 100 μL of the given antifungal agent (final inoculum size of 5 × 102 to 2.5 × 103 CFU/mL). Then, the plates were cultured at 35ºC for 48 h. The resistance breakpoint categories of 5-flucytosine, itraconazole, and fluconazole were used according to the CLSI M27-A3 criteria (25) while ketoconazole and clotrimazole were based on previous investigations (26), as shown in Table 1. The minimal inhibitory concentrations (MICs) were determined as the lowest concentration causing at least 80% growth inhibition compared to control wells not containing the antifungal agent. The MICs for amphotericin B and nystatin were identified as the minimum concentration yielding a complete growth inhibition.
3.4. RNA Extraction
Total cellular RNA was isolated from C. albicans at mid-exponential growth phase (OD600 = 0.8) using the hot phenol method (27). Briefly, cells were collected by centrifugation at 4000 rpm for 5 min at room temperature, the supernatant was removed, and then the pellets were spun down a second time to remove all liquid. Next, cell pellets were resuspended in 800 μL of AE buffer (50 mm sodium acetate, 10 mm EDTA, pH 5.2). Then, 80 μL of 10% SDS and 880 μL of pre-warmed phenol (pH 5.2) were added and the samples were vortexed thoroughly. The tubes were transferred to a 65ºC water bath and vortexed for 5 s once every 1 min for 5 min. Lysates were transferred and kept in a dry ice/ethanol bath for 2 - 3 min, followed by centrifugation at 13,200 rpm for 10 min at room temperature.
The aqueous layers were carefully transferred to new microcentrifuge tubes and an equal volume of phenol (pH 5.2): chloroform: isoamyl alcohol (25: 24: 1) was added. Then, samples were mixed by vortexing and centrifuged again at 13,200 rpm for 10 min at room temperature. The aqueous layers were then carefully transferred to new microcentrifuge tubes. RNA was precipitated by the addition of 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volume of chilled 100% ethanol, followed by incubation at -20ºC overnight. Precipitated RNA was collected by centrifugation at 13,200 rpm for 15 min at 4ºC. The entire supernatant was decanted; the RNA pellet was washed with 1 mL of chilled 75% ethanol and centrifuged again. The pellet containing RNA was air-dried in a hood for 5 - 10 min and then resuspended in 100 μL of DEPC water; then, it was treated with RNase-free DNase (ThermoFisher, USA) to prevent genomic DNA contamination. The quantity and quality of RNA were assessed by the measurement of the OD 260/280 absorption ratio and running the denaturing formaldehyde agarose gel.
3.5. Quantitative Real-Time PCR
The cDNA was synthesized using 2 μg of total RNA according to the manufacturer’s instruction of the PrimeScriptTM RT Reagent kit with gDNA Eraser (Takara, Japan). Gene-specific PCR primers for RTA2, CDR1, CDR2, ERG11, FLU1, and MDR1 genes and 18S rRNA were designed using Primer Premier 5.0 (Table 2). The 18S rRNA was included as an internal reference gene. The qRT-PCR analysis was performed using the 7500 Real-time PCR System (Applied Biosystems, USA) and the SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) kit (Takara, Japan). All samples contained 10 μL of 2X SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (including TaKaRa Ex Taq® HS, dNTP Mixture, Mg2+, Tli RNaseH, SYBR® Green I), 0.8 μL of forward and reverse primers, 1 μL of cDNA template, 0.4 μL of ROX Reference Dye, and 7.2 μL of nuclease-free water.
| No. | Primer Name | Sequence (5’ - 3’) |
|---|---|---|
| 1 | CaERG11a-F | CAAGAAGATCATAACTCAAT |
| CaERG11a-R | CAGAACACTGAATCGAAAGA | |
| 2 | CaERG11b-F | TTTGGTGGTGGTAGACATAGAT |
| CaERG11b-R | TAATCAGGGTCAGGCACTTT | |
| 3 | CaCDR1-F | GATTCTCAAACTGCCTGGTC |
| CaCDR1-R | CCAAAATAAGCCGTTCTTCCAC | |
| 4 | CaCDR2-F | AAAAAGGTGGAAGAACGGC |
| CaCDR2-R | TTGGCATGAGATCCTGGTG | |
| 5 | CaMDR1-F | TGCGTCAAGAACAGGTTITC |
| CaMDR1-R | AAGCAGTAGTAGCAGCACC | |
| 6 | CaFLU1-F | TGGATAGTCCCCGTCATTGG |
| CaFLU1-R | GGCAAAAAGTGGGAAAACAGC | |
| 7 | CaRTA2F | ATGTCAAATCGGTAAGAGGTC |
| CaRTA2R | AGCCAATTCTGCCACTCTAT | |
| 8 | 18S RNA-F | TCTTTCTTGATTTTGTGGGTGG |
| 18S RNA-R | TCGATAGTCCCTCTAAGAAGTG |
The PCR conditions were as follows: 40 cycles of denaturation for 5 s at 95ºC, annealing for 34 s at 60ºC for 18S rRNA, MDR1, CDR2, CDR1, FLU1, RTA2, and ERG11, and a melt curve step (from 60ºC, gradually increasing at 0.5ºC/s to 95ºC, with acquisition data every 1 s). Fluorescent data were collected during the annealing step and analyzed with ABI software. The threshold cycle (ΔCT) value was obtained by calculating the difference between the CT values of the target gene and the normalizer (18S rRNA). The mean mRNA levels for each gene were calculated from at least three independent biological replicates. The expression levels of target genes were analyzed according to the 2-ΔΔCt method (28). Differences between resistant and susceptible groups were analyzed with independent sample t-test.
3.6. Statistical Analysis
The results are expressed as means ± standard deviation (SD). Comparisons between the sensitive and resistant groups were made using independent-samples t-tests by SPSS 13.0 software. The differences were considered statistically significant when the P values were less than 0.05.
4. Results
4.1. Antifungal Susceptibility
As described in Table 3, all fluconazole-resistant strains were susceptible to 5-flucytosine. Moreover, 25 (67.74%) fluconazole-resistant isolates were also resistant to ketoconazole, 28 (70.97%) were resistant to itraconazole, and 23 (64.52%) were resistant to clotrimazole. Interestingly, the fluconazole-resistant isolates, which were associated with the mutation of ERG11, exhibited a higher MIC for nystatin, while the isolates that were not associated with the mutation of ERG11 were cross-resistant to other azoles.
| Isolate | FlZ | MIC, μg/mL | Amino Acid Change (s) in Erg11p | |||||
|---|---|---|---|---|---|---|---|---|
| KETO | ITR | CLOT | 5-FC | NYS | B | |||
| 56388 | 64 | 8 | 4 | 2 | 2 | 16 | 4 | A114S Y257H |
| 49372 | 64 | 0.5 | 16 | 0.5 | 2 | 8 | 1 | A114S Y257H |
| 49922 | 64 | 0.5 | 4 | 0.25 | 0.125 | 8 | 1 | A114S Y257H |
| 56539 | 64 | 0.25 | 0.5 | 0.25 | 1 | 8 | 1 | A114S Y257H |
| 57451 | 64 | 1 | 2 | 4 | 0.25 | 4 | 4 | A114S Y257H |
| 57800 | 64 | 0.0625 | 0.5 | 0.625 | 0.125 | 4 | 2 | A114S Y257H |
| 58181 | 64 | 1 | 4 | 0.25 | 0.125 | 4 | 2 | A114S Y257H |
| 59161 | 64 | 0.25 | 2 | 0.25 | 0.125 | 4 | 2 | A114S Y257H |
| 59182 | 64 | 0.5 | 1 | 2 | 4 | 8 | 1 | A114S Y257H |
| 56472 | 64 | 16 | 2 | 0.25 | 0.25 | 0.125 | 0.5 | D116E K128T Y132H G465S |
| 59690 | 64 | 16 | 8 | 0.25 | 0.5 | 8 | 2 | D116E K128T Y132H G465S |
| 51527 | 64 | 1 | 0.5 | 0.25 | 4 | 2 | 2 | G450E Y132H |
| 58614 | 64 | 0.0313 | 0.0313 | 0.25 | 0.125 | 16 | 4 | Y132H G450E |
| 56392 | 64 | 2 | 16 | 0.5 | 0.125 | 8 | 4 | Y132H G488E |
| 56292 | 64 | 8 | 16 | 1 | 0.125 | 4 | 1 | A114S Y257H |
| 54535 | 64 | 0.5 | 0.125 | 0.125 | 0.125 | 2 | 2 | -a |
| 49340 | 64 | 8 | 4 | 2 | 1 | 2 | 1 | -a |
| 49345 | 64 | 0.5 | 16 | 0.5 | 0.25 | 4 | 1 | -a |
| 56350 | 64 | 16 | 16 | 2 | 0.125 | 1 | 4 | -a |
| 56525 | 64 | 8 | 16 | 1 | 0.125 | 4 | 4 | -a |
| 57598 | 64 | 8 | 16 | 0.5 | 0.25 | 4 | 4 | -a |
| 49312 | 64 | 16 | 0.25 | 0.25 | 0.125 | 2 | 1 | D116E |
| 49477 | 64 | 0.5 | 16 | 0.25 | 0.125 | 4 | 1 | D116E |
| 56452 | 64 | 16 | 4 | 0.0625 | 0.25 | 0.125 | 2 | D116E |
| 56477 | 64 | 16 | 16 | 0.25 | 0.125 | 4 | 4 | D116E |
| 56507 | 64 | 8 | 16 | 0.5 | 0.5 | 4 | 4 | D116E |
| 56533 | 64 | 16 | 16 | 0.5 | 1 | 8 | 4 | D116E |
| 59145 | 64 | 16 | 1 | 1 | 0.25 | 4 | 2 | D116E |
| 57464 | 64 | 16 | 1 | 0.25 | 0.25 | 2 | 4 | D116E E266D |
| 59537 | 64 | 0.0625 | 0.125 | 0.625 | 0.125 | 4 | 0.5 | D116E E266D V488I |
| 59407 | 64 | 1 | 0.25 | 0.25 | 0.25 | 4 | 2 | D116E V488I |
| 57856 | 64 | 0.125 | 1 | 0.25 | 0.125 | 4 | 4 | D11DE E266D V488I |
| 56682 | 64 | 16 | 16 | 1 | 0.125 | 4 | 1 | D225H E266D |
| 56517 | 64 | 16 | 16 | 2 | 0.125 | 4 | 2 | E266D |
| 56689 | 64 | 0.0313 | 0.125 | 0.625 | 0.125 | 4 | 2 | E266D |
| 56262 | 64 | 1 | 16 | 1 | 8 | 4 | 4 | E266D V437I V488I |
| 59679 | 64 | 16 | 0.5 | 2 | 0.25 | 4 | 2 | E266D V488I |
| 56214 | 64 | 4 | 16 | 0.5 | 0.25 | 8 | 4 | E266D V488I |
| 57442 | 64 | 0.125 | 0.5 | 1 | 0.25 | 4 | 0.5 | K342R |
| 55475 | 64 | 64 | 0.25 | 0.25 | 0.125 | 0.125 | 4 | V437I |
Abbreviations: B, amphotericin B; CLOT, clotrimazole; FLZ, fluconazole; ITZ, itraconazole; KETO, ketoconazole; NYS, nystain.
aIsolates with no missense mutation in ERG11.
4.2. Expression of ERG11
The melting peaks and curves of the ERG11 amplicon indicated that there were no non-specific amplification products or primer dimers produced during the reactions. Further, a single band representing the PCR product with the expected length on an agarose gel confirmed the specificity of the PCR (data not shown). Gene expression levels of ERG11 from 40 fluconazole-resistant and -susceptible isolates were analyzed by the real-time PCR. The mean ΔCt values of the ERG11 gene in the fluconazole-resistant group and the susceptible group were 16.80 ± 0.18 and 17.16 ± 0.26, respectively, with no statistically significant difference (P = 0.25).
4.3. The Expression of Efflux Pumps
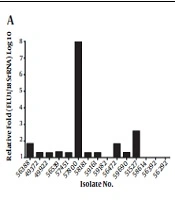
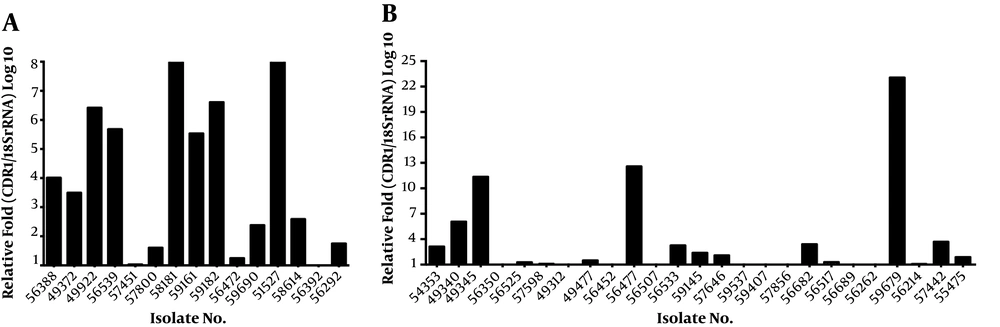
The CDR1 expression level was 3.68 folds higher in the fluconazole-resistant group than in the fluconazole-susceptible group (Figure 1). The mean ΔCt values of CDR1 expression were 15.92 ± 0.28 and 16.80 ± 0.24 in the resistant and susceptible groups, respectively. The expression of CDR1 was significantly different between the fluconazole-susceptible and resistant groups (P = 0.0193). Among the 40 fluconazole-resistant isolates, isolate 59679 displayed the highest level of CDR1 gene expression (23.07 folds) (Figure 1B). In our previous work, we found that only 36.73% (19/49) of the fluconazole-resistant isolates were associated with the mutation in the ERG11 gene. In this study, we further analyzed the differences between isolates related and unrelated to the mutation in the ERG11 gene and found that CDR1 expression was altered in 80.00% of the fluconazole-resistant isolates containing a mutant variant of ERG11, and that the expression level of CDR1 was more than 1.5-fold higher in these isolates (Figure 1A).
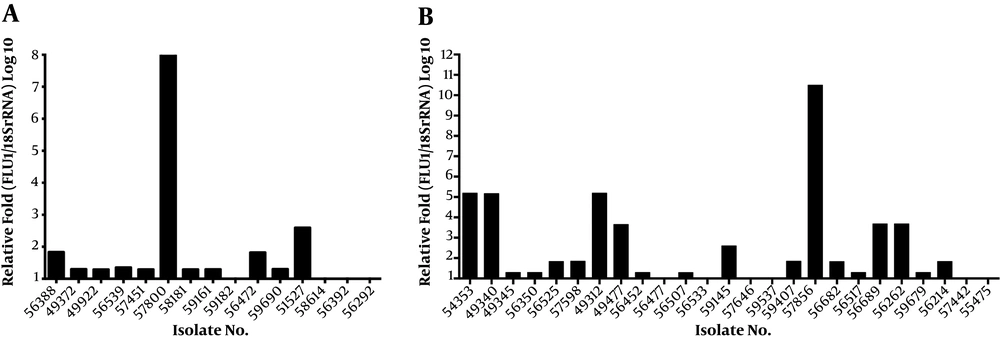
Among the 40 fluconazole-resistant isolates, 31 (77.50%) isolates exhibited 1.29 to 10.50-fold upregulation of the FLU1 gene (Figure 2). The mean ΔCt values of the FLU1 gene were 17.64 ± 0.16 and 18.39 ± 0.15, respectively, in the resistant and susceptible groups. The difference in FLU1 gene expression was 0.0010 between fluconazole-resistant and susceptible groups, which was statistically significant (P = 0.0010). Compared to the susceptible isolates, the expression levels of FLU1 were not upregulated in most resistant isolates, which carried a mutant variant of ERG11. The mean ΔCt values of the CDR2 and MDR1 genes were 15.96 ± 0.25 and 21.51 ± 0.31, respectively, in the resistant group and 16.27 ± 0.45 and 21.72 ± 0.17, respectively, in the susceptible group. The mRNA expression levels of CDR2 and MDR1 were not statistically different between fluconazole-resistant and susceptible groups (P = 0.55 and 0.56, respectively).
4.4. Expression of RTA2
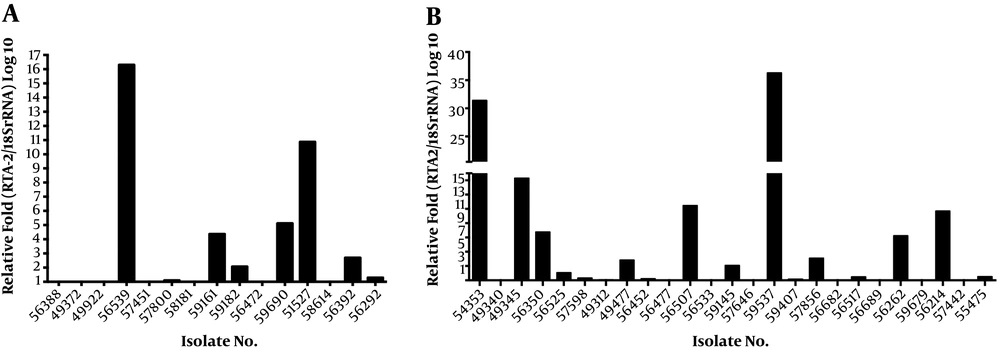
Among 40 fluconazole-resistant isolates, 19 (47.5%) isolates exhibited more than 1.5-fold upregulation in the RTA2 gene when compared to the fluconazole susceptible isolates, and the mean RTA2 gene expression in resistant isolates was 4.91 folds higher (Figure 3). The mean ΔCt values of the RTA2 gene were 16.08 (SD = 0.28) and 17.19 (SD = 0.20), respectively, in the resistant and susceptible groups, which indicated that RTA2 gene expression was significantly different between these two groups (P = 0.0017).
5. Discussion
Azole is a widely used antifungal agent for the treatment of both superficial mucosal and deep disseminated Candida infections. However, the widespread application of azoles and the structural similarity of these molecules have resulted in the development of cross-resistance to various members of this class of drugs (29). Previous studies demonstrated that fluconazole resistance was correlated with cross-resistant to other azoles (30). Recently, researchers found that about 74% of fluconazole-resistant strains were also resistant to ketoconazole and itraconazole (31). Our present study demonstrated that most fluconazole-resistant isolates were cross-resistant to other azoles.
Resistance to a variety of drugs is defined as multidrug resistance (MDR). In infectious bacteria, such as Mycobacterium tuberculosis, the emergence of MDR is caused by a series of point mutations in different target genes (32). A similar process can be found in fungal infections, as well. Azoles inhibit the enzyme lanosterol 14α-demethylase, which is encoded by the ERG11 gene. A mutation in the ERG11 gene or its overexpression may affect the enzyme’s affinity for drugs, resulting in resistance (10). However, in our previous study, 63.27% (31/49) of the fluconazole-resistant isolates were not associated with point mutations in the ERG11 gene, indicating that other factors are involved in azole resistance in these strains. In this study, we further investigated the relationship between ERG11 expression/mutation and fluconazole resistance. The overexpression of the ERG11 gene may increase the production of drug target enzymes to an extent exceeding the inhibitory capacity of antifungal drugs, which may, in turn, contribute to fluconazole resistance. However, the role of overexpression of ERG11 in fluconazole resistance remains enigmatic. Some studies have suggested that the overexpression of ERG11 is not significantly related to fluconazole resistance in C. albicans (33). Similar to previous reports, we did not find any significant difference in the expression of ERG11 between multi-azole susceptible and resistant strains in this study.
In Candida, another important mechanism of drug resistance is associated with efflux transporters. In this study, we aimed to elucidate the role of efflux transporters in the development of fluconazole resistance. As previously reported, the overexpression of the CDR gene is one of the most predominant mechanisms of MDR in azole-resistant Candida clinical isolates. Prior studies found that a drug efflux pump-encoding CDR gene contributed to the development of the cross-resistance phenotype in C. glabrata strains (34). Mdr1p, encoded by the MDR gene, is a member of the MFS transporters, and is able to pump several structurally unrelated compounds out of the cell, including fluconazole. The expression levels of CDR1, CDR2, and MDR1 genes increased in most clinical isolates with fluconazole MICs of ≥ 64 μg/mL, while the disruption of these genes resulted in hypersensitivity to azoles (34). Furthermore, the increased expression of CDR1, CDR2, and MDR1 genes was a major contributor to azoles resistance in clinical isolates (21, 35-38). In this study, our results suggested that the CDR1 gene was upregulated in fluconazole-resistant isolates. However, unlike most reports, no correlation was observed between MDR and overexpression of CDR2 and MDR1 in this study.
FLU1, encoding Flu1p, is a multidrug efflux transporter implicated in mycophenolic acid resistance. Similar to CDR, FLU1 was discovered in a genomic library screened for the complementation of fluconazole hyper-susceptibility in the Pdr5 (ABC transporter gene) mutant Saccharomyces cerevisiae isolates (20). When the FLU1 gene was heterologously expressed in S. cerevisiae, it mediated both fluconazole and cycloheximide resistance (20). The deletion of FLU1 leads to insignificant changes in susceptibility to fluconazole. However, the deletion of FLU1 in a strain based on the disruption of other genes encoding multidrug efflux pumps (such as CDR1, CDR2, and MDR) may cause increased susceptibility to several azole derivatives (20). Similarly, the gene product of FLU1 has been found to mimic Tpo1 from S. cerevisiae, which is a primary plasma membrane polyamine efflux transporter (39). Further study found that FLU1 is able to pump Histatin 5 out of the cell and reduce the toxicity of Histatin 5 in C. albicans (40). However, the overexpression of FLU1 is generally uncommon among clinical resistant isolates of C. albicans. In most studies, changes in the expression level of FLU1 were not significant between azole-resistant and susceptible C. albicans isolates (21, 37). Interestingly, our findings showed that upregulation of FLU1 was one of the dominant mechanisms in the fluconazole-resistant isolates analyzed in this study.
Several lines of evidence suggest that fluconazole resistance may involve many unknown mechanisms that have yet to be elucidated. Recently, studies found that calcium signaling plays an important role in the development of drug resistance and it may be a target for overcoming drug resistance (41, 42). A novel gene, RTA2, which mediates calcineurin-dependent resistance to azoles, was found to contribute to the development of fluconazole resistance (23, 43). The knockdown of RTA2 leads to higher susceptibility of C. albicans to fluconazole. Conversely, ectopic expression of RTA2 resulted in markedly decreased fluconazole efficacy in mice with systemic Candida infections (44). Furthermore, previous studies found that the RTA2 gene was over-expressed in both laboratory and clinical resistant strains (45). Consistently, the RTA2 expression levels elevated in our present study.
Our previous results showed that only 36.73% of fluconazole-resistant strains were associated with point mutations in ERG11. Interestingly, we also found that 75.0% of the fluconazole-resistant isolates exhibited the overexpression of FLU1 gene, 62.5% were associated with upregulation of CDR1 (more than 1.5-fold expression), and 45% showed high levels of RTA2 expression. In addition, we found that 10% of the fluconazole-resistant isolates were associated simultaneously with ERG11 mutation and overexpression of RTA2, CDR1, and FLU1 genes.
These results indicate that multiple genes are associated with fluconazole resistance. Additionally, the upregulation of CDR1 was a major mechanism in fluconazole-resistant isolates having point mutations in ERG11. Interestingly, we found that more than 90% of the fluconazole-resistant isolates with A114S and Y132H ERG11 variants exhibited the upregulated expression of CDR1. It seems that the specific ERG11 mutant variants A114S and Y132H may be associated with CDR1 overexpression. However, further studies need to investigate the relationship between CDR1 expression and ERG11 variants A114S and Y132H. For the isolates that were not related to mutant variants of ERG11, the overexpression of FLU1 and RTA2 was a major contributor to drug resistance.
5.1. Conclusions
In conclusion, we demonstrated that the overexpression of FLU1 and RTA2 was correlated with azole resistance; this finding had not been reported previously in clinical isolates of C. albicans. Taken together, this study provides useful information for the treatment of candidiasis and indicates that clinicians should be cautious of cross-resistance within this class of antifungal drugs, especially for the treatment of patients with prior azoles prophylaxis or patients at high risk of C. albicans infections. Moreover, fluconazole resistance in C. albicans is a multifactorial phenomenon with complicated mechanisms. Therefore, it is important to notice that most of these mechanisms are frequently combined in a single isolate to contribute to a step-by-step acquisition of fluconazole resistance.