1. Background
Hospital infections are a global health problem associated with a variety of factors. The causes of these infections are various among different countries. Since the 1980s, Gram-positive bacteria, especially Staphylococcus aureus, has been considered as the main cause of hospital infections (1). The biofilm formation by S. aureus and its resistance to antimicrobial agents have made this organism a major problem for hospitals and medical staff (2). The S. aureus causes a wide range of infections, including bacteremia, septicemia, and pneumonia, as well as skin, soft tissue and bone infections (3). Staphylococcus aureus is a frequent cause of hospital- and community-associated infections on a global scale (4). However, S. aureus can also cause a series of other diseases, including endocarditis or osteomyelitis. Often, S. aureus infection can proceed to septicemia and become life-threatening. Furthermore, S. aureus is a frequent cause of biofilm-associated infections, in particular those developing on indwelling medical devices (5). Finally, S. aureus can cause food poisoning (3).
Some S. aureus virulence factors such as alpha-toxin and phenol-soluble modulins (PSMs) encode on the core genome, which is virtually produced by all strains. Alpha-PSM peptides of S. aureus are toxins that play a role in infection and neutrophil lysis after phagocytosis. It is an important mechanism in the pathogenesis of highly invasive strains of S. aureus (6). The alpha 1 PSM and alpha 2 PSM peptides have antibacterial activity by lysing competitive bacteria (7). The PSM activates the host immune system by regulating the production level of lymphokine (8). Phenol-soluble modulins, also have a wide range of functional activities, including supporting biofilm formation during S. aureus infections (9). During S. aureus growth in biofilms, bacteria become tolerant to concentrations of antimicrobials that could eliminate single-cell bacteria, making biofilm infections particularly difficult to eradicate (10).
Studies show the multi-faceted role of PSMs in biofilm development: it helps in making bacterial biofilms’ structure by its surfactant activity but on the other hand, its expression can also lead to biofilm dispersal, i.e. the detachment of cells or cellular clusters from biofilms, which is a key strategy leading to the systemic dissemination of biofilm infection (11). In regard to the importance of biofilm as a key factor in survival and antimicrobial resistance of S. aureus strains, the prevalence of biofilm-producing S. aureus and identification of genes involved in biofilm formation is highly significant, especially in clinical samples. The discovery that PSMs have been recognized as key players in staphylococcal pathogenesis, prompted us to determine expression of the PSM A as a first gene of PSM class of peptides (9).
2. Objectives
There is no information about the expression of PSM A gene in clinical isolates of biofilm-producing S. aureus in Iran. In the present study, we report, for the first time, the prevalence and expression of PSM A gene in biofilm-producing S. aureus clinical isolates in Iran.
3. Methods
3.1. Samples and Identification
Samples were collected from different wards of 5th Azar Hospital, Gorgan, Golestan province, Iran from different sources (wound, CSF fluid, urine culture, abscess, pleural fluid and ascites fluid) from September 2018 to March 2019. The samples were examined for S. aureus using conventional bacteriological methods such as culture on manitol salt agar, Gram staining, other biochemical tests (catalase, oxidase and coagulase) and manitol fermentation or DNase test if necessary (12). For all biochemical tests, the standard strain of S. aureus ATCC 25923 was used as positive control.
3.2. Biofilm Formation
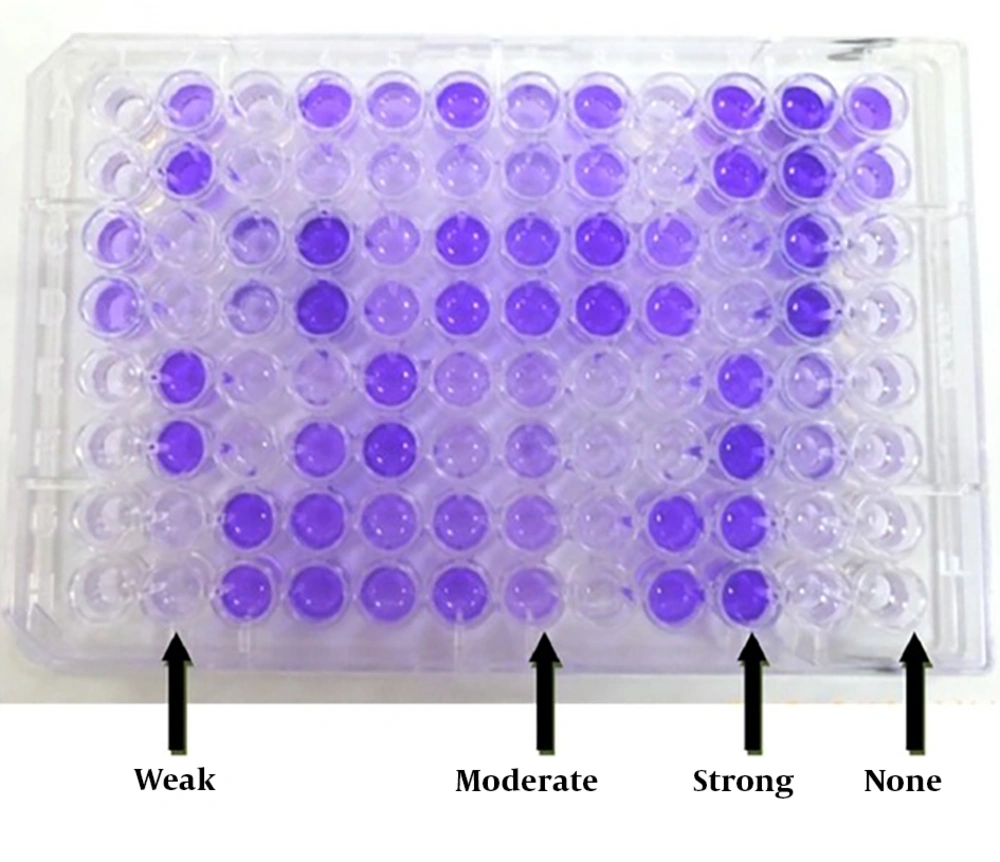
Biofilm study on S. aureus (Biofilm Power): The ability of biofilm formation in the isolates was examined by microtiter plate (crystal violet) (13, 14) as following: A 24-hour culture of each bacterial isolate was prepared and inoculated in Mueller Hinton agar medium, TSB medium containing 1% glucose and incubated at 37°C. When the opacity of the tubes were equal to 0.5 McFarland standard (0.08% - 0.13% absorbance at a wavelength 625 nm) (12), 200 μL of suspension was inoculated into a 96-well polystyrene microplate. As a negative control, 200 μL of TSB medium with 1% glucose without bacteria was added to other wells and S. aureus ATCC 35556 (the strain of strong biofilm producer) was used as a positive control (15). To view biofilms, 200 μL of crystalline violet 2% was added to each well for 5 minutes.
The excess stain was discarded and then washed three times with PBS. To remove the crystal from the bacteria and biofilm, 200 μL of ethanol-acetone (20% to 80%) was added to each well. After 30 minutes, the optical absorption of each well was measured by an ELISA reader at a wavelength of 570 nm. A semi-quantitative study of biofilm formation using cut-off calculations was carried out in accordance with the following formula (16, 17). The mean OD of negative controls + three times the standard deviation was considered as a cut- off point. To ensure the correctness of the work for the isolates studied, the absorbance of each isolate was examined three times. The method of calculating the quotient amount for each group is presented in Table 1 and Figure 1.
| Biofilm Formation Ability | Calculate the Cut-Offa | Results of Mean (Maximum OD Absorption) |
|---|---|---|
| Strong | OD > 4 × ODc | OD > 1.2 |
| Moderate | 2 × ODc < OD ≤ 4 × ODc | 0.7 < OD ≤ 1.2 |
| Weak | ODc < OD ≤ 2 × ODc | 0.3 < OD ≤ 0.6 |
| None | OD < cut off | OD ≤ 0.3 |
aCut-off: average of OD negative control + (3 × SD of negative control).
3.3. Evaluation of PSM A Expression in Biofilm-Producing Staphylococcus aureus Isolates
3.3.1. Extraction and Purification of RNA
RNA extracted from S. aureus biofilm for studying on S. aureus PSM A gene expression. RNA extraction from bacterial biofilm was performed using the RNX-Plus kit (CinnaGene Co.), these steps were as follows: For total RNA isolation RNX-Plus, a guanidine/phenol solution which contains guanidine thiocyanate and phenol, was used to extract the entire RNA from homogeneous samples. The solution was prepared in a volume of 25 mL from the CinnaGene company and maintained at 2 - 8°C. The bacterial isolates were each grown for 24 hours at 37°C in Mueller Hinton agar. The medium was poured and washed three times with distilled water to remove non-adherence free cells and replaced by fresh preheated medium every day .For extraction, in addition to the solution, chloroform, isopropanol, 75% ethanol and water treated with DEPC were required. After the disruption, 1 mL of RNX-Plus solution was added to the lysed product and mixed well for 5 to 10 seconds and incubated at room temperature for 5 minutes. After that, 200 μL of chloroform was added and washed with vigorous shaking for 15 seconds (not vortex). The micro tube was incubated for 4 minutes at 5ºC. The sample was centrifuged for 15 minutes at 12000°C and at 4°C.
3.3.2. Evaluation of Extracted RNA Quality
The quality and quantity of extracted RNA was evaluated using electrophoresis and agarose gel 1.2% for qualitative evaluation and nanotubes or biophotometers for quantitative ones.
3.3.3. Expression of PSM A
To determine the amount of gene expression in extracted RNA, the real-time PCR of the SYBR Green bark was used. To perform this RealQ Plus 2x Master Mix Green Kit was applied. Real-time PCR was achieved by an ABI Prism 7300 Applied Biosystems thermocycler. The primers were confirmed at the NCBI site, using the Blastas shown in Table 2. Genomic amplification was performed by using QuantiFast SYBR Green Master Mix (Qiagen, Hilden, Germany) (as described in Table 3). The reactions were mixed appropriately into the PCR stacks and cDNA sample (1 - 10 ng) added to each well containing a reaction mixture. The amplifications were performed on a ABI Prism 7300 Applied Biosystems device (Applied Biosystems, Foster, CA, USA) and thermal cycling protocol (as shown in Table 4).
| Name | Sequence (5 to 3) | Product Size (bp) | Reference |
|---|---|---|---|
| PSM A (F) | CTTTCACATGGGTATCATTGCAGG | 266 | (8) |
| PSM A (R) | CAATAGCCATCGTTTTGTCCTCCT |
| Component | Volume/ Reaction | Final Concentration |
|---|---|---|
| 2x QuantiFast SYBR Green Master Mix | 12.5 µL | 1 x |
| Primer F | 1 µL | 1 µM |
| Primer R | 1 µL | 1 µM |
| Template of cDNA | 4 µL | 5 ng (1 - 10 ng) |
| RNase-free water | 6.5 µL | - |
| Total reaction volume | 25 µL | - |
| Step | Time | Temperature |
|---|---|---|
| PCR initial activations | 15 min | 95°C |
| Denaturation | 15 s | 95°C |
| Combined annealing/extension | 60 s | 60°C |
| Number of cycles | 35 | |
3.4. Statistical Analysis
Descriptive quantitative variables were computed by calculating central indexes and dispersion and plotting, and qualitative variables were computed by calculating the percentage of frequency. Also, information obtained from the samples and the results of the evaluations was entered into SPSS V. 16 software and analyzed by ANOVA and chi-square. In all cases a significant level of P < 0.05 was considered.
4. Results
4.1. Isolation and Identification of Staphylococcus aureus
A total of 60 strains of S. aureus were isolated from 1800 clinical isolates. Of 60 S. aureus isolates, 27 (47%) were isolated from men and 33 (53%) were isolated from women. The mean age of patients with S. aureus bacteremia was 37.8 years and the highest rate of S. aureus carriers was in the age group of 15 - 25 years (40%). There was no statistically significant correlation (P = 0.03) between the mean age of patients with S. aureus infection.
4.2. Quantitative Biofilm Production Test
Of the 60 S. aureus clinical isolates included in this study, 47 strains (78.3%) were identified as biofilm producer and the others 13 (21.7%) were negative for biofilm formation. The biofilm-forming strains were isolated from urine (n = 12), blood (n = 5), cerebrospinal fluid (n = 8), wound (n = 17) and abscess (n = 5) specimens. The microtiter plates assay results showed that 30 (50%) isolates were strong biofilm producers and 17 (28.3%) isolates were weak biofilm producers and 13 isolates (21.7%) were found as non-adhesive. No obvious correlation was observed (P < 0.05) between source of bacteria and sex of patients, and potential of biofilm formation in strains.
4.3. Determination of PSM A Gene
All the primers used in the study showed specificity with a single band. After real-time PCR testing to detect PSM gene it was found that 100% of the strains were positive for biofilms and PSM A genes. The results of phenotypic and genotypic tests of biofilm were closely related to each other and the expression of PSM A gene was 80%. In addition, the results showed that 100% of strains were biofilm producers and the PSM A gene was present in 47 (78.3%) strains.
5. Discussion
Staphylococcus aureus is one of the important bacteria with the capability to stick and produce biofilms on external surfaces which cause community-acquired infections. Increasing antibiotic resistance among biofilm producers in hospital settings makes the therapy of staphylococcal infection an international challenge (13). Several studies showed the biofilm formation and genetics characteristics of different isolates of S. aureus (19-22). Consistent with the result of our study, Nourbakhsh and Momtaz reported the 86% of S. aureus isolates isolated from hospital infections in Shahrekord (the capital city of Chaharmahal and Bakhtiari province, Iran) were able to form biofilms (23). Arya et al. also reported the same results. They isolated 52 strains of S. aureus from blood culture of volunteers. In this study, 87% of strains isolated from blood culture were positive for biofilms (24). Christensen et al. showed that 48.5% of the clinical strains of S. aureus were able to produce biofilms (25). Similar to the current research, in a study published in 2007, Cafiso et al. reported that 57.5% of S. aureus strains isolated from in-patient medical equipment were biofilm producers (26).
In this study, according to quantitative evaluation of biofilm production, 30 isolates (50%) as sticky strains, 17 strains (28.3%) as weak adhesive strains and 13 strains (21.7%) as non-adhesive strains were considered. Mirzaee et al. in 2014 reported that 46% of clinical isolates of methicillin resistant S. aureus (MRSA) were strong producer of biofilm on microtiter tissue culture plates (21). Croes et al. in 2009 proposed that strong biofilm formation in S. aureus could not be attributed to a specific accessory gene regulator (agr) genotype, as suggested previously (27). Eftekhar and Dadaei in 2011 showed that 53.3% of MRSA isolates had the potential to form biofilm by colony morphology of which, 75% carried the ica operon. Weak biofilm production was observed in the microtitre plate assay by 57.8%, of which 53.8% harbored the ica operon. However, about 70% of biofilm non-producers also carried the ica operon (28).
Namvar et al. detected the intercellular adhesion gene cluster (ica) in clinical S. aureus isolates. Their results showed that in the quantitative biofilm assay, 58% produced biofilm while 42% isolates did not exhibit this property (29). In present study, 47 (78.3%) strains were identified as biofilm- producing strains and 13 (21.7%) strains were negative for biofilm. According to the results of studies from all different parts of the world, it seems that the ability to produce biofilms among strains isolated from patients is very close to the present study, which indicates the cloning of patients with these strains has a high prevalence in Iran.
Based on our knowledge, expression of some genes is responsible for adhesion and biofilm formation in bacteria. Quantification of the gene expression involved in biofilm formation by S. aureus strains has been investigated in several studies (21, 22). In S. aureus, all PSMs have biofilm-structuring activities through their physico-chemical properties (11). In this study, the prevalence and expression of PSM A gene in clinical isolates of biofilm producing S. aureus was evaluated. The PSM A genes were present in 78.3% of all biofilm-producing isolates. Consistent with our result, in the Fursova et al. study, the PSM A gene was detected in 81.6% of the isolates (30).
Given that biofilm production and pathophysiology of S. aureus infection are multifactorial and controlled by various genetic factors, a complete review of the genes involved in the process of biofilm formation is needed. The high prevalence of biofilm strains of S. aureus in patients is a warning and a serious health risk to the community. The introduction of these bacteria into the genitourinary system will become a major problem that will make it really difficult to eliminate and eradicate. Such a large-scale study in Iran is absolutely essential for screening and access to patient information.
5.1. Conclusions
The high prevalence of biofilm producing S. aureus isolates could pose a major health challenge with serious consequences for hospitalized patients. Therefore, it is crucial to disinfect and sterilize hospital surfaces and equipment effectively to minimize the risk of contamination and spread of bacteria in the hospital settings.