1. Background
Klebsiella pneumoniae is a bacterium of enterobacterium Klebsiella, and an important human pathogen (1). Urinary tract infection, pneumonia, and bacteremia are common symptoms of K. pneumoniae infection (2). The mortality rate of infection with K. pneumoniae is up to 60% and approximately 41 to 50% in bloodstream infections, which has aroused particular concerns (3-5). There are increasing reports of meningitis, osteomyelitis, necrotizing fasciitis, endophthalmitis, and other severe infections caused by hypervirulent variants of K. pneumoniae (6, 7). Besides, K. pneumoniae is associated with multidrug resistance, especially to carbapenems and third-generation cephalosporins, posing a huge threat to public health worldwide (8, 9).
In China, the infection rate of K. pneumoniae is second to that of Escherichia coli, and developing a user-friendly, rapid method to screen K. pneumoniae is of great significance (10-12). At present, many methods are used to detect K. pneumoniae, such as bacterial culture and biochemical identification, polymerase chain reaction (PCR), and Loop-mediated isothermal amplification (LAMP) (13, 14). However, the identification of K. pneumoniae in clinical practice is primarily based on bacterial culture and biochemical identification combined with PCR (15, 16). The method is easy to operate and even it is regarded as the gold standard in practical application (17). However, the drawbacks of this method are prominent. The enrichment of K. pneumoniae and the genomic extraction are still needed before PCR and LAMP assay (18). Moreover, a set of PCR equipment is essential, making it expensive and unavailable in hospitals, especially in some developing countries. Thus, the method is not suitable for rapid detection.
Recently, a LAMP assay has been reported to monitor K. pneumoniae (19). Loop-mediated isothermal amplification is a novel technique, which can amplify DNA with high specificity, sensitivity, and rapidity (20). The total time for detection is about one hour without the need for much equipment such as PCR. The results can be observed by agarose gel electrophoresis or judged by the naked eye using SYBR green I (20). However, the enrichment of bacteria and a bacteria genomic DNA kit are necessary before LAMP assay. Some special samples, such as sputum and blood, are difficult for enriching. Therefore, currently, neither LAMP nor PCR is suitable for rapid and accurate detection of K. pneumoniae.
Immune magnetic bead (IMB) separation technology is a new biological detection method for enrichment of bacterial cells (21). In this method, a specific antibody is conjugated with magnetic beads. The IMB is able to enrich bacteria isolated from the environment under the action of an outside magnetic field (22, 23). In this study, three monoclonal antibody (mAbs) specific to K. pneumoniae are prepared. Then, protein A/G magnetic beads are conjugated with mAb 1E6 to generate immuno-magnetic beads. In this study, an immunocapture assay employing immunomagnetic beads combined with LAMP, named immunocapture loop-mediated isothermal amplification (IC-LAMP), is developed for the first time to detect K. pneumoniae. The IC-LAMP was more sensitive and less expensive than PCR.
2. Objectives
In this work, we developed a simple, rapid, and visual detection method by employing the IC-LAMP based on the ureR-1 gene. The sensitivity of IC-LAMP was 4 CFU mL-1 and had no cross-reaction with other microbial strains. Besides, the accuracy of IC-LAMP was further verified by examining 39 clinical isolates. The IC-LAMP was efficient in pathogenic cell enrichment, free of template plasmid or genome extraction, with a one-step procedure. The IC-LAMP developed here can be used as a primary screening method supplementary to traditional detection methods to improve the diagnosis of K. pneumoniae infection.
3. Methods
3.1. Strains
The study used 27 strains, including 20 K. pneumoniae strains and seven other strains (Supplementary File Appendix 1). Besides, 39 clinical isolates (20 K. pneumoniae strains and 19 non-K. pneumoniae strains) were collected from patients in the First Affiliated Hospital, Kunming University of Science and Technology, and used for IC-LAMP assay (Supplementary File Appendix 2).
3.2. Production of pAbs and mAbs Against K. pneumoniae
Polyclonal antibodies (pAbs) and monoclonal antibodies (mAbs) against K. pneumoniae were produced as described by Zhang et al. (24, 25). Klebsiella pneumoniae was inactivated by treatment at 85°C for 1 hour, and then mixed with the Freund’s complete adjuvant and injected into mice. The mice were injected two more times with 108 inactivated K. pneumoniae in incomplete Freund’s adjuvant, two weeks and four weeks after the first injection. Finally, the spleen cells of the immunized mice were collected and washed three times with RPMI 1640 (Gibco, USA). The spleen cells were, then, fused with SP2/0 cells (stored at our laboratory). For the culture of the fused cells, we used the selection medium containing 20% FBS (Gibco, USA) and 1% hypoxanthine-aminopterin-thymidine (Sigma, USA). Ten days later, the supernatant of the medium was replaced by fresh RPMI 1640 containing 20% FBS and 1% hypoxanthine-thymidine (Sigma, USA). The K. pneumoniae-specific antibodies were screened by enzyme-linked immunosorbent assay (ELISA) in the supernatants of hybridoma.
3.3. Preparation and Purification of Ascites
Three cycles of subcloning were successfully performed to prepare anti-K. pneumoniae positive hybridoma cells. Three hybridomas were washed twice with RPMI 1640, respectively, and then each was injected to one female Balb/c mice with 106 hybridoma cells. One week later, the ascites were collected. Then, Protein A-Sepharose (GE Healthcare, Chicago, IL, USA) was used to purify all ascites and the ascites were titered by ELISA. The immunoglobulin concentration of the three antibodies was determined by the BCA (bicinchoninic acid assay ) assay.
3.4. Antibody Isotype Determination
The immunoglobulin isotypes of the produced mAbs were determined using a mouse mAb isotyping kit (SouthernBiotech, SBA clonotyping System-HRP Kit), according to the manufacturer’s instructions.
3.5. Titration of Ascites
After the ascites were purified, the titer of each antibody was determined by ELISA. In brief, 100 µL of 108 CFU mL-1K. pneumoniae (inactivated) was added to each well and incubated at 37°C for 2 hours. Then, all wells were washed three times with phosphate-buffered saline (PBS) containing 0.05% tween-20, blocked with 200 µL of blocking buffer (5% skimmed milk in PBS-T) for 1 hour at 37°C, and washed as described earlier. Next, 100 µL of the three antibodies with different dilutions (from 1:200 to 1: 409600) was added to each well and maintained for 2 hour at 37°C. Then, each well was washed as described earlier and incubated with the goat anti-mouse IgG (H + L) horseradish peroxidase (HRP) (GenScript, USA) diluted for the final concentration of 0.25 µg mL-1. After washing, the soluble TMB substrate solution (TIANGEN, China) was added to each well, followed by incubation for 15 minutes at 37°C. Finally, the reaction was terminated by the stop buffer (2 M H2SO4), and the optical density of each well was measured at 450 nm.
3.6. Western Blot
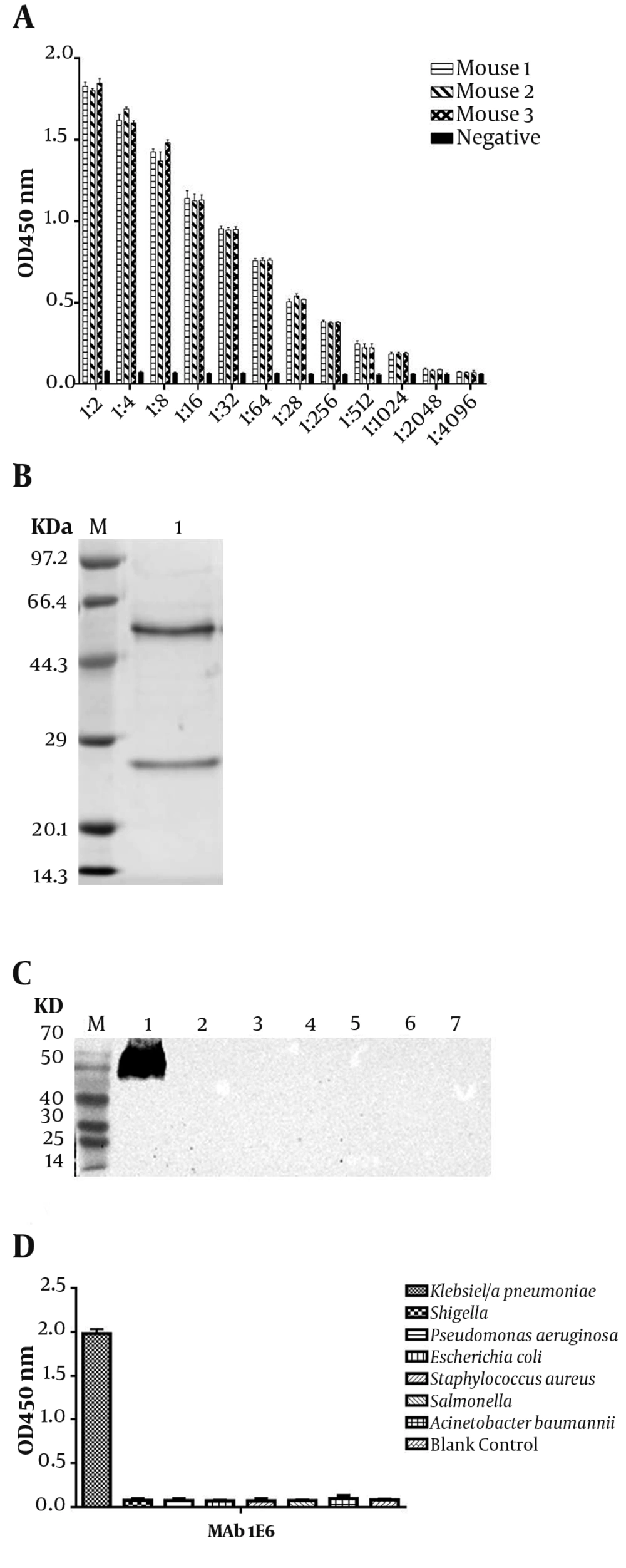
The characteristics of mAb 1E6 were determined by Western Blot. In brief, seven common strains (K. pneumoniae, Shigella, Staphylococcus aureus, Pseudomonas aeruginosa, E. coli, Acinetobacter baumannii, and Salmonella) were cultured in LB liquid medium at 37°C in a shaking incubator set at 160 rpm. Then, all cells were harvested and suspended in PBS and inactivate. Next, the lysate of each bacterium was subjected to sodium dodecyl sulphate-poly acrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose (NC) membrane. The NC membrane was blocked with the blocking buffer. The ascites were diluted for the final concentration of 1.5 µg mL-1 with blocking buffer and incubated for 2 hours at 37°C. Next, the goat anti-mouse IgG (H + L) HRP (diluted as above) was added, followed by incubation for 1 hour at 37°C. Finally, the reactivity of mAb 1E6 was detected by a Western Blot kit (BioBest, Anhui, China).
3.7. Primer Design
The primers of IC-LAMP and PCR were designed according to the ureR-1 gene (GenBank accession no. 11847803), which was predicted as a specific gene of K. pneumoniae by local and online BLAST. As shown in Figure 1, the primers were selected by its amplification efficiency for the target gene.
3.8. The Normal LAMP Reaction
The LAMP reaction was performed in a 25 µL volume containing two pairs of primers (K-F3, K-B3, K-FIP, and K-BIP), 15 µL of the 2 × isothermal master mix, genomic DNA, and distilled water. The LAMP reaction was performed at 65°C for 30 minutes and terminated at 80°C after 2 minutes. The products were analyzed by electrophoresis.
3.9. Preparation of IMB
The IMB was prepared as described by Zhang et al. (23). In brief, 10 mg of protein A/G magnetic beads (Biotool, United States) were washed three times with binding buffer (150 mM NaCl, 50 mM Tris, 0.1% TritionX-100, pH 7.5). Then, mAb 1E6 was diluted in the binding buffer for the final concentration of 50 µg mL-1. The prepared protein A/G magnetic beads were mixed with 300 µg of mAb 1E6 coated at 37°C for 30 minutes in a shaking incubator set at 100 rpm. After magnetic separation, the rest of mAb 1E6 in tubes was removed and washed three times with PBS. After washing, the immune magnetic beads (IMBs) were diluted in PBS and stored at 4°C for further use.
3.10. Standard IC-LAMP Assay
Klebsiella pneumoniae cells in different samples were captured with IMBs at 37°C for 0.5 hour in a shaking incubator set at 100 rpm. Then, the samples were removed and the IMBs were washed three times with washing buffer. The mixture (breads-mAb-K. pneumoniae) was then lysed with 10 µL of solution A (1 mM EDTA, 125 mM NaOH, 0.1 % tween-20) for 10 minutes at 65°C and terminated by 10 µL of solution B (10 mM Tris, 125 mM HCl). The lysates were used for the LAMP reaction.
3.11. Specificity and Sensitivity of LAMP and IC-LAMP
Seven common pathogenic bacteria were cultured and used for the specificity of LAMP and IC-LAMP. At the same time, the sensitivity of LAMP and IC-LAMP assays was also identified using serial dilutions of bacteria ranging from 4 × 108 to 4 × 100 CFU mL-1 prepared with mouse blood.
3.12. Detection of K. pneumoniae in Clinical Isolates by IC-LAMP Assay
Klebsiella pneumoniaeitalic> is a strong pathogenic bacteria, mainly causing pneumonia, sepsis, and urinary tract infections. Klebsiella pneumoniae screening is a necessary component of thorough inspections in hospitals. We tested the direct detection of K. pneumoniae in patients’ clinical isolates. The IC-LAMP assay was applied for the detection of 39 clinical isolates collected from hospital settings. All clinical isolates were mixed with PBS. Then, IMBs were added to capture K. pneumoniae cells in mixtures at 37°C for 0.5 hour. When bacterial cells were adsorbed, all tubes were washed thoroughly with wash buffer. The captured cells were used for IC-LAMP assay.
3.13. Isolation and Identification of K. pneumoniae
In a laminar flow bench, all sputum samples were suitably diluted, applied to the gallbladder and sulfur milk agar plate (DHL) (Nissui Pharmaceutical, Tokyo, Japan), and cultured for 12 hours at 37°C. After inoculation, the colonies of K. pneumoniae on DHL were pale pink, large and uplift, smooth and moist, and mucus, and adjacent colonies were easy to integrate. Five typical or suspected colonies were picked and punctured in semi-solid agar, and inoculated to the nutrient agar slant. Then, the Gram’s staining and oxidase test were applied to identify the cells. The verification of the cells was done by the VITEK 32 biochemical identification system. After physiological and biochemical identification, the genomes of these suspicious bacteria were extracted using a Bacterial Genomic DNA kit (Zomanbio, China). The extracted DNA was amplified by PCR.
4. Results
4.1. Generation and Characterization of Specific mAbs Against K. pneumoniae
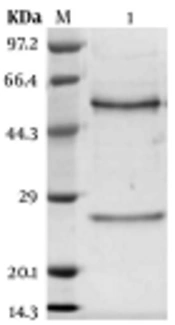
Three Balb/c mice were immunized with 108K. pneumoniae after inactivation, which were emulsified by complete or incomplete Freund’s adjuvant. After the fourth immunization, the titer of sera was tested by ELISA (Figure 2A). Then, three mAbs were generated and designated as 2F1, 1E6, and 4E6, after cell fusion and subcloning. The titer, subtypes, and KD value of 2F1, 1E6, and 4E6 were determined in our previous study (26). In the next step, mAb 1E6, which had the highest affinity, was purified and confirmed by SDS-PAGE (Figure 2B). The target protein of mAb 1E6 was identified by immunoprecipitation and mass spectrometry. The results of mass spectrometry showed that mAb 1E6 recognized outer membrane protein C (Supplementary File Appendix 3). The reactivity and specificity of mAb 1E6 were evaluated by Western Blot and ELISA. The results of Western Blot and ELISA showed that mAb 1E6 could recognize only K. pneumoniae (Figure 2C lane 1 and Figure 2D) and Shigella, E. coli, A. baumannii, S. aureus, P. aeruginosa, and Salmonella were not recognized (Figure 2C lanes 2 - 7 and Figure 2D).
Characterization of anti-Klebsiella pneumoniae sera and ascites. (A) the titer of mice antiserum; (B) the SDS-PAGE analysis of purified mAb 1E6; (C) Western Blot analysis of K. pneumoniae ascites 1E6; Lane 1 represents K. pneumoniae lysates; lanes 2 to 7 represent Shigella, E. coli, A. baumannii, S. aureus, P. aeruginosa, and Salmonella, respectively; (D) the ELISA of the specificity of mAb 1E6.
4.2. Optimization Operation
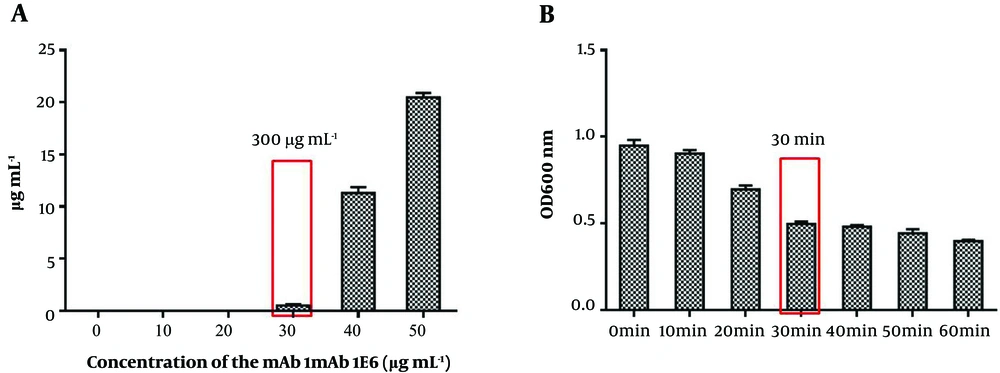
To develop a rapid and sensitive detection method, we carefully optimized each step of IMB preparation. First, the capacity of magnetic beads was calculated with mAb 1E6 serial dilutions and then the remaining concentration of the liquid was measured. Figure 3A shows that the capacity of magnetic beads was 300 µg mL-1. Besides, the time of K. pneumoniae capturing by IMBs was optimized from 10 minutes to 60 minutes. As shown in Figure 3B, as the incubation time increased from 10 minutes to 30 minutes, the value of OD600 decreased significantly. However, when the incubation time raised to 60 minutes, no significant differences were observed in OD600. Hence, the optimal time for IMBs to capture K. pneumoniae cells was 30 minutes. The binding efficiency of IMBs was determined by preparing serial dilutions and plate counting. Plate counting was used to calculate the initial number of cells in the dilutions of 106, 104, and 102 CFU mL-1 and the remaining cells in the solutions after capturing by IMBs (three-time replication). The results showed that the average binding efficiency was 88% (Supplementary File Appendix 4).
4.3. Sensitivity and Specificity of IC-LAMP
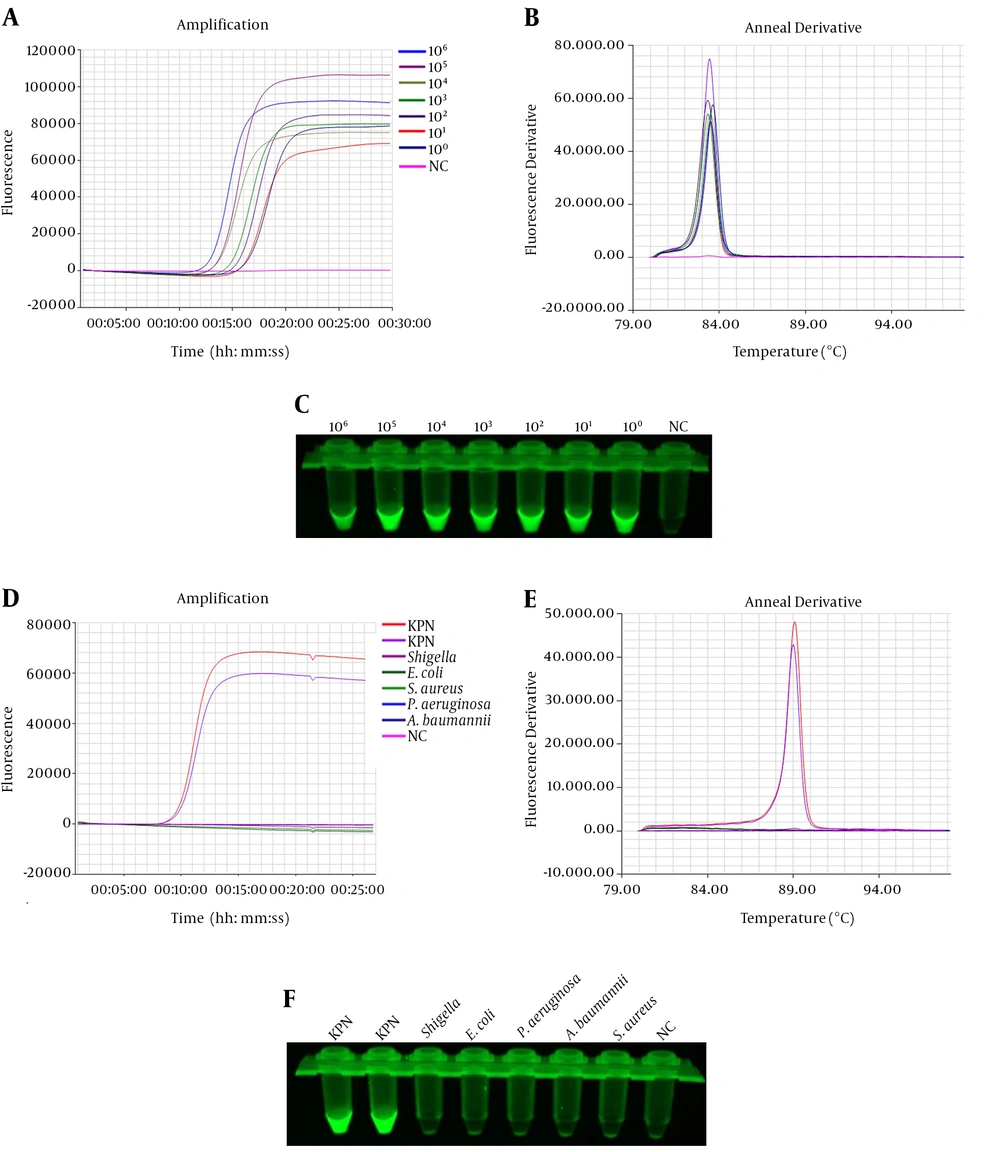
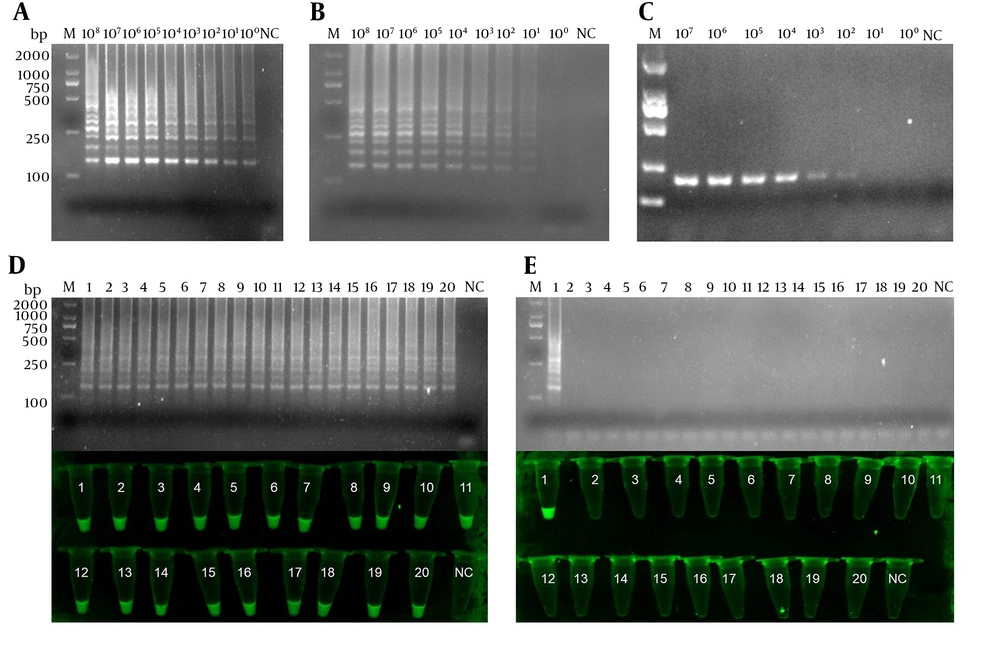
The sensitivity of IC-LAMP was verified strictly in serial dilutions of 4 × 108 to 4 × 100 CFU mL-1. The results of the sensitivity test revealed that the detection limit of IC-LAMP was up to 4 × 100 CFU mL-1 (Figure 4A and C). Figure 4D and F show that the amplification curves and green fluorescence were obvious in the reaction containing K. pneumoniae. On the contrary, there were no amplification curves and green fluorescence in the experiments of the other five bacteria. Compared with the sensitivity of normal LAMP and PCR, we can clearly observe that the detection limit of IC-LAMP was several times higher than that of normal LAMP and PCR in blood samples (Figure 5A - C). That is, LAMP combined with magnetic immunocapture improved the sensitivity of detection to some certain extent.
Sensitivity and specificity of the IC-LAMP assays. Amplification (A) and annealing (B) curves of the sensitivity of IC-LAMP assay; (C) sensitivity of the IC-LAMP assays based on SYBR Green I detection under UV light; Amplification (D) and annealing (E) curves of the specificity of IC-LAMP assay; (F) specificity of the IC-LAMP assays based on SYBR Green I detection under UV light.
The comparison of sensitivity between IC-LAMP, LAMP, and PCR for examining 39 clinical isolates. The sensitivity of IC-LAMP (A), LAMP (B), and PCR (C); (D) the results of 20 positive clinical isolates analyzed by electrophoresis and based on SYBR green I detection under UV light; (E) the results of 19 negative clinical isolates analyzed by electrophoresis and based on SYBR green I detection under UV light.
4.4. Detection of K. pneumoniae in Clinical Isolates by IC-LAMP
The accuracy of IC-LAMP was verified further. Therefore, 39 clinical isolates collected from patients in the hospital were subjected to IC-LAMP. The IC-LAMP revealed 20 positive K. pneumoniae products (Figure 5D) and 19 negative products (Figure 5E). Meanwhile, the 39 clinical isolates were further identified by the gold standard test. The obtained results were consistent with the IC-LAMP test results, demonstrating that IC-LAMP developed in this study could be used to quickly, accurately, and specifically detect K. pneumoniae in clinical isolates (Table 1).
| Results | Methods | ||
|---|---|---|---|
| IC-LAMP | PCR | Gold Standard Testa | |
| Positive | 20 | 18 | 20 |
| Negative | 19 | 21 | 19 |
| Total | 39 | 39 | 39 |
Comparison of IC-LAMP, PCR, and Traditional Biochemical Identification Methods
5. Discussion
The high mortality rate of infection with K. pneumonia necessitates developing a rapid and sensitive detection method. K. pneumoniae is a common human pathogen causing some serious infectious diseases, such as pneumonia, urinary tract infection, and sepsis (27). Besides, a significant treatment challenge has appeared with the extensive use of antibiotics, leading K. pneumoniae to quickly obtain resistance to all kinds of antibiotics (28, 29). In many hospitals, patients infected with K. pneumoniae encounter a grim situation of difficult drug selection and even no drug availability, contributing to the high mortality rate of K. pneumoniae infections (28, 30). Thus, prior to treatment, a rapid and sensitive detection method can save much valuable time for patients and doctors.
The traditional culture-based and biochemical confirmation methods are time-consuming with low sensitivity. They cannot detect K. pneumoniae in a short time especially in hospital settings. The PCR and LAMP are the common methods used in the detection of the bacterium. These methods with the characteristics of high sensitivity and few steps have been applied to the detection of various pathogens, including fungi, parasites, viruses, and bacteria (31, 32). However, they are still dependent on bacterium enrichment and genomic extraction, which would take several hours. For many special samples including blood, sputum, and urine samples, the steps of enrichment are difficult to conduct.
In this study, the LAMP combined with magnetic immunocapture (IC-LAMP) was developed to detect K. pneumoniae in artificial samples and clinical isolates. Prior to designing the experiment, we found many studies in the literature reporting PCR and LAMP as the main techniques to detect K. pneumoniae. However, there was no report of IC-LAMP use for the detection of K. pneumoniae. Several studies reported the detection of parasites, fungi, bacteria, and viruses by IC-PCR (33, 34). Many studies used polyclonal antibodies (pAbs) for this purpose (33). Although the process of preparation is relatively easy, simple, and rapid, there are obvious disadvantages for pAbs. Additionally, there may be differences between different batches of pAbs, making it difficult for use in large scales. The pAbs also may produce nonspecific antibodies. Overall, pAbs are not suitable for detection studies on a large scale. On the contrary, mAbs have many advantages that pAbs did not have. The mAbs have chemical structures that can be selected to have a defined specificity for a particular analyte (target molecule). They can be produced in unlimited quantities.
Considering the above-mentioned reasons, K. pneumoniae was used as an antigen to generate mAbs. After cell fusion, three stable anti-K. pneumoniae antibody-positive hybridoma cells were prepared successfully. Then, the antibody was combined with LAMP for IC-LAMP. This study is the first to combine mAbs with magnetic beads based on LAMP to develop the IC-LAMP. Compared with the normal PCR and LAMP, the IC-LAMP combines the specificity of both antibodies and LAMP, and it is more effective in the enrichment of pathogenic cells. Without the need for cell culture and genome or plasmid extraction, the IC-LAMP is more rapid and convenient than other detection methods (13, 14). Besides, the detection limit of K. pneumoniae in the established IC-LAMP was 4 CFU mL-1 that was higher than that of PCR and traditional detection methods (35, 36). Moreover, the specificity of IC-LAMP was checked by other similar bacteria. The practical application of IC-LMAP for K. pneumoniae detection was also confirmed by testing 39 clinical isolates. The results of examining 39 clinical isolates by IC-LAMP were consistent with the gold standard test results. Therefore, IC-LAMP developed in this study could detect K. pneumoniae accurately and quickly.
5.1. Conclusions
A novel, specific, reliable method for the detection of K. pneumoniae strains was developed based on LAMP combined with magnetic immunocapture assay (IC-LAMP). By targeting the ureR-1 gene, two pairs of primers were designed and applied to IC-LAMP. Based on the specificity of antibody and LAMP, the IC-LAMP can be more convenient, rapid, specific, and sensitive than other detection methods. Moreover, the practical application of IC-LAMP was successful in detecting 39 clinical isolates. Therefore, the novel IC-LAMP assay could be used as a potential tool for facilitating the monitoring of K. pneumoniae strains in a variety of clinical isolates and guiding for clinical medication and timely patient treatment.
5.2. Limitations and Suggestions
It should be noted that in this study, the IC-LAMP efficacy was confirmed only by examining 39 clinical isolates (20 K. pneumoniae strains and 19 non-K. pneumoniae strains). The 20 K. pneumoniae strains were all drug-resistant, and could contain the genes kpc, okp, tem, shv, act, ctx-m, oxa, and so on. However, we could not make sure that the IC-LAMP could cover all drug-resistant bacterial strains due to the limited number of clinical samples. It was very difficult for us to collect all drug-resistant strains of K. pneumoniae. Thus, we hope that the IC-LAMP developed here can be applied to detect more isolates of K. pneumoniae to confirm its efficacy. Besides, we screened mAb 1E6 against the outer membrane protein C of K. pneumoniae. We hope that mAb 1E6 is a useful tool and makes more contributions to the diagnosis of K. pneumoniae, as well as treatment of its infection.