1. Background
Pseudomonas aeruginosa is an opportunistic pathogen that is common in nature and causes both exogenous and endogenous infections. Endogenous infections are often the result of colonization. In particular, colonization of the gastrointestinal tract in neutropenic patients may result in bacteraemia and pneumonia that might be occurred in intubated patients as a result of the endotracheal colonization. Other infections, such as burn injury, conjunctivitis, keratitis, and urinary tract infection, caused by P. aeruginosa, are mostly exogenous (1). The P. aeruginosa has natural resistance to most antibiotic classes and many antibacterial agents can develop resistance even during treatment (2). The development resistance occur through chromosomal mutations or as a result of horizontal transitions of resistance genes via plasmids, transposons and integrons. Acquired resistance is caused by efflux pump, decrease of outer membrane permeability and beta lactamase and carbapenemase formation (1). Epidemiological studies show that infections caused by resistant to P. aeruginosa increase morbidity, mortality, surgical intervention requirement, surgical intervention, length of stay in hospital, and cost of treatment (2).
Wide spectrum penicillins, 3rd and 4th generation cephalosporins, carbapenems, monobactams, fluoroquinolones, aminoglycosides and various antibiotics such as colistin are active against P. aeruginosa. However, mutational resistance may develop to all of antipseudomonal antibiotics (1). Carbapenems are a new class of beta-lactam antibiotics that have been used since 1980 in the treatment of infections caused by P. aeruginosa. Carbapenem resistance that develops along with the use of carbapenems is common throughout the world. This resistance depends on the loss of the P. aeruginosa outer membrane protein OprD, the excretion of the drug via efflux pumps, and the release of a carbapenem-hydrolyzing enzyme (2-4).
Metallo-beta-lactamases (MBL) that hydrolyze carbapenems are found in Group 3 according to the Bush-Jacoby-Medeiros functional classification and in Class B according to the Ambler molecular classification (5, 6). The most commonly encountered transferable MBL are IMP and VIM. There are also the SPM-, GIM-, SIM-, KHM-, NDM-, AIM-, DIM-, SMB-, TMB-, and FIM-type enzymes (7, 8). Today, 33 of the 55 known IMP variants and 24 of the 46 known VIM variants have been detected in P. aeruginosa. However, blaVIM-2 is the most frequently detected MBL type in P. aeruginosa (8). Some phenotypic methods based on the disc diffusion method have been proposed for rapid detection of MBL production. The activity of these enzymes is zinc-dependent. They are inhibited by ethylenediaminetetraacetic acid (EDTA) or mercaptopropionic acid (MPA). These tests are based on the principle that the activity of MBL enzymes is inhibited by metal chelators such as EDTA and 2-MPA. The double disc synergy test, the combined disc test (CDT), and the MBL E-Test can be shown as an example (9).
Molecular tests have a high specificity in detecting the presence of MBL. The polymerase chain reaction (PCR) is considered the easiest and fastest method. However, the requirement of DNA primers with high specificity, inability to distinguish the variants, and failure to identify new variants are the disadvantages of this method. Other molecular methods used in MBL detection are isoelectric focusing, polyacrylamide gel electrophoresis, and nucleotide sequence analysis (9).
2. Objectives
The aim of the study is to determine MBL production by phenotypic and genotypic methods in the isolates of imipenem- and/or meropenem-resistant P. aeruginosa isolated from clinical samples sent to the Medical Microbiology Laboratory from the inpatient services and outpatient clinics in our hospital.
3. Methods
3.1. Identification of Isolates and Antibiotic Susceptibility Tests
The 58 patients who were intermediate susceptible/resistant to imipenem and/or meropenem according to the antibiotic susceptibility test (AST) results were included in this study. The Vitek 2 GN identification kit (BioMerieux, France) was used for identification of species. These isolates were subjected to the Kirby-Bauer disc diffusion test for imipenem and meropenem. ASTs were performed using the Vitek AST cards (BioMerieux, France) for isolates intermediate susceptible or resistant to any of imipenem or meropenem. Thus, the minimal inhibitory concentrations were determined. The results of the AST were categorized as susceptible, intermediate susceptible, and resistant according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria. Moreover, the Gradient Test (the E-test, Biomerieux, France) was performed for imipenem and meropenem in order to confirm carbapenem resistance; and to determine the minimum inhibitory concentration (MIC) value, the P. aeruginosa strain ATCC 27853 was used as a standard strain (10).
3.2. Phenotypic Identification of MBL
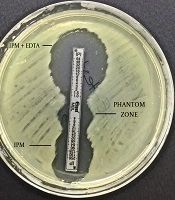
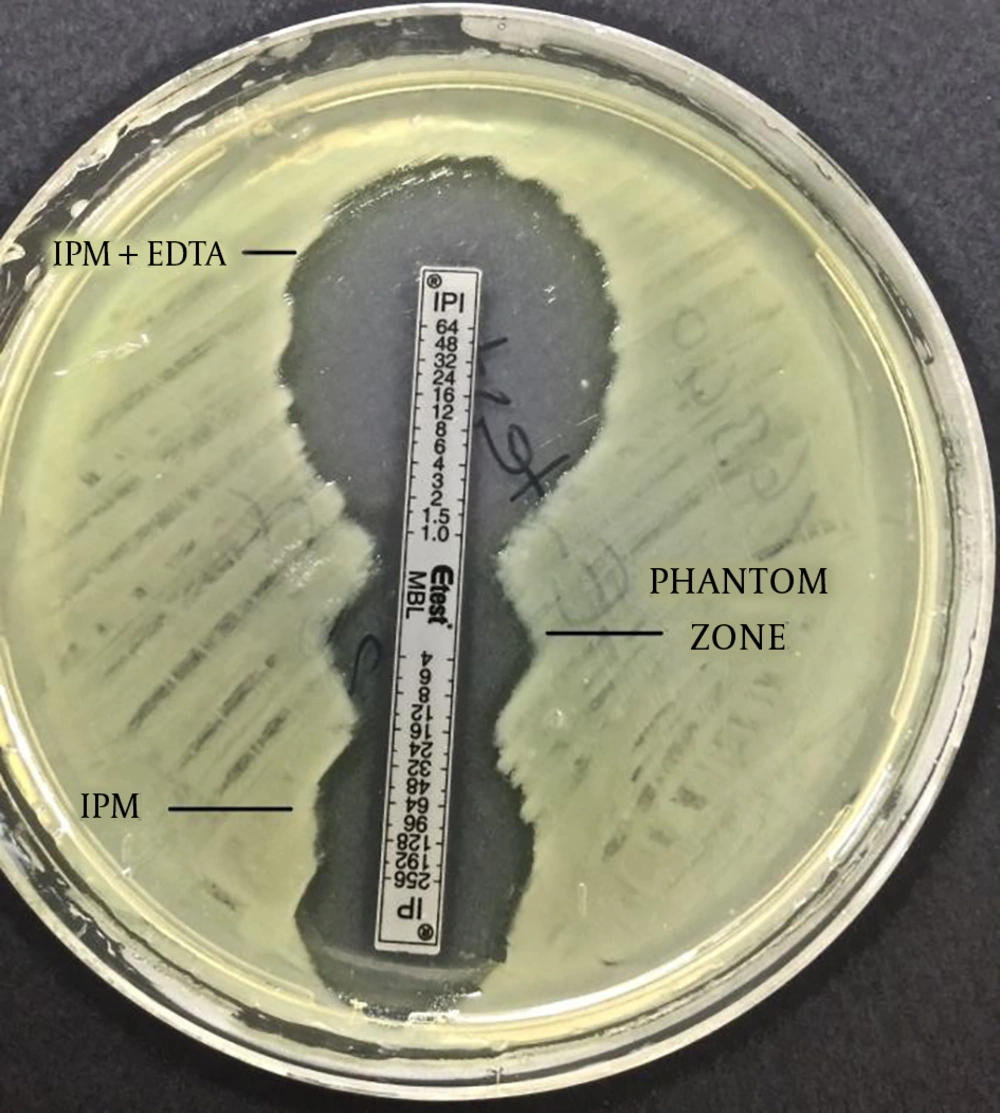
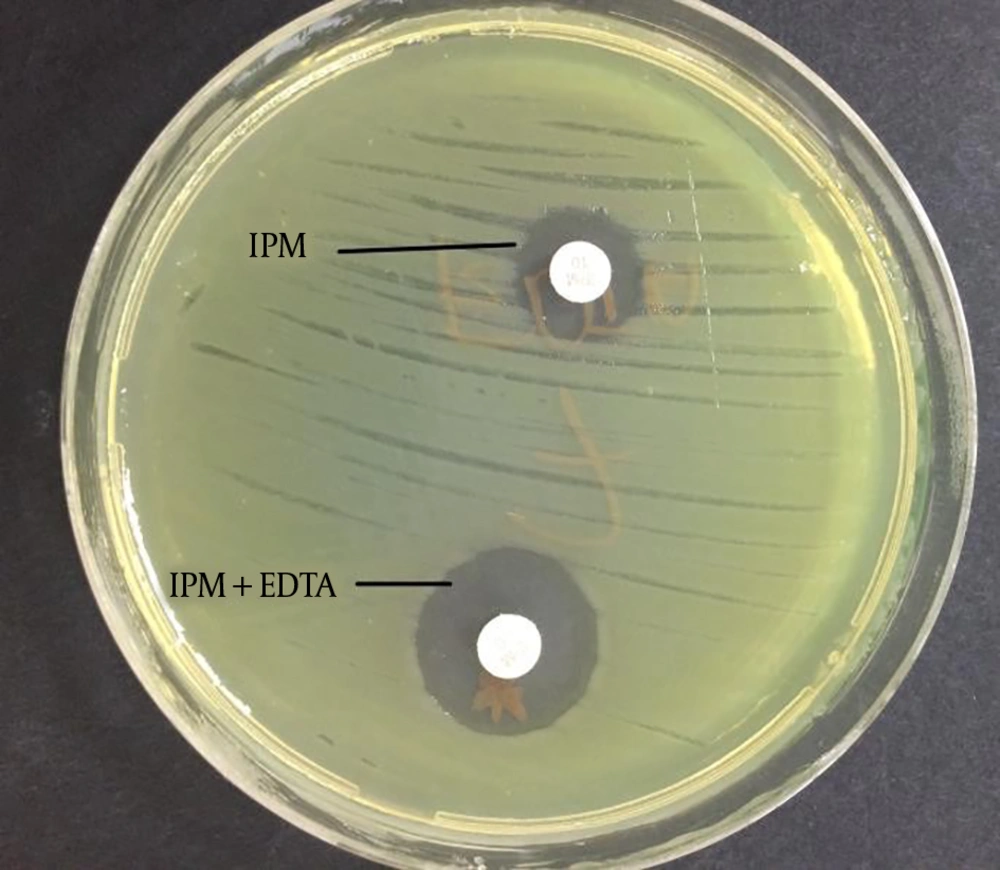
The imipenem-EDTA CDT and the MBL E-test were performed to phenotypically demonstrate the production of MBL. The bacterial suspensions were prepared in sterile saline (0.9%) and the turbidity of suspension was adjusted to 0.5 Mc Farland standards in dansitometer machine (DEN-1B, Biosan). Prepeared suspensions were homogeneously spread on the surface of Mueller Hinton agar medium with the aid of a swab. For the CDT, IPM (10 μg) and IPM-EDTA (750 μg) discs were placed in the plate. Before the test, 10 μL of 0.5 M EDTA solution was instilled into one of the IPM discs with the aid of a pipette. Then, this disc was expected to absorb the solution. Zone diameters were measured after incubation at 36 ± 1ºC for 16 - 18 hours. The result was considered positive if the inhibition zone diameter around the IPM disc with the addition of 0.5 M EDTA was at least 7 mm greater than the inhibition zone diameter around the IPM disc without the addition of 0.5 M EDTA (11, 12). For the MBL E-test, imipenem/imipenem EDTA E-Test (BioMerieux, France) strips were placed after the plate surface dried. Plates were assessed after incubation at 37°C for 16 - 18 hours. A ≥ 8 fold decrease in the MIC of imipenem in the presence of EDTA was considered as a positive test indicative of MBL (9).
3.3. Molecular Analysis
The boiling method was used for DNA isolation from clinical isolates which were cultured on 5% sheep blood agar. Firstly, the colonies were vortexed in 500 μL TE buffer (10 mMTris, 0.5 mM EDTA). The suspension was heated at 95°C for 10 minutes. After it was centrifuged at 14,000 rpm for 4 minutes, 400 μL of the supernatant containing bacterial DNA was transferred into a new microcentrifuge tube. It was stored at -20°C until PCR procedures (3). The presence of the blaIMP-1, blaIMP-2, blaVIM-1, blaVIM-2, blaGIM-1, and blaSPM genes were investigated by the PCR. The primers used in the reaction are shown in Table 1. For all gene regions, the PCR amplification of each sample was performed using 50 μL reaction volumes. The reaction mixture contained 5 μL 10X PCR buffer, 2 μmol/μL MgCl2, 0.2 μmol/μL dNTP mix, 0.25 pmol/μL of each primer (sense and antisense), 1.25U of Taq DNA polymerases, and 5 μL of sample DNA. The amplification conditions for the samples in the “thermal cycler” (Eppendorf, Mastercycler, Germany) included an initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 20 seconds, annealing at 53°C for 45 seconds, extension at 72°C for 30 seconds, and final extension at 70°C for 6 minutes (13-15). The PCR products were stained with 0.5 μg/mL ethidium bromide after 1% agarose gel electrophoresis. Subsequently, they were viewed with an ultraviolet (UV) transilluminator.
| Primers | Oligonucleotide Sequences (5’-3’) | Base Pair Length (bp) | Reference |
|---|---|---|---|
| IMP-1F | ATGAGCAAGTTATCTGTATTC | 741 | 10 |
| IMP-1R | TTAGTTGCTTGGTTTTGATGG | ||
| IMP-2F | ATGAAGAAATTATTTGTTTTATG | 741 | 10 |
| IMP-2R | TTAGTTACTTGGCTGTGATG | ||
| VIM-1F | AGTGGTGAGTATCCGACAG | 799 | 10 |
| VIM-1R | ATGAAAGTGCGTGGAGAC | ||
| VIM-2F | ATGTTCAAACTTTTGAGTAAG | 801 | 10 |
| VIM-2R | CTACTCAACGACTGAGCG | ||
| SPM-F | CCTACAATCTAACGGCGACC | 649 | 11 |
| SPM-R | TCGCCGTGTCCAGGTATAAC | ||
| GIM-1F | AGAACCTTGACCGAACGCAG | 599 | 11 |
| GIM-1R | ACTCATGACTCCTCACGAGG |
aReference sequences: LC169578.1, CP008857.1.
3.4. DNA Sequencing Analysis of the PCR Products
In order to verify the samples that were positive for the blaIMP-1, blaIMP-2, blaVIM-1, blaVIM-2, blaGIM-1, and blaSPM genes by the PCR and the “Cycle Sequence” PCR of the sense and antisense strands was performed on the amplified gene products using “the BigDye Terminator V. 3.1 Cycle Sequencing Kit” (Applied Biosystems, Foster City, CA, USA) according to the instructions of the manufacturer. After the purification process, the reaction products were subjected to electrophoresis using an automated DNA sequence analysis device “ABI PRISM 3130XL Genetic Analyzer” (Applied Biosystems, Foster City, CA, USA). Thus, DNA sequence analysis data were obtained chromatographically.
4. Results
Of the 58 isolates that were collected during the study period, 15 (25.8%) were from internal medicine clinics, 12 (20.6%) were from intensive care unit (ICU), 10 (17.2%) were from surgical clinics, 6 (10.3%) were from medical ICU, 6 (10.3%) were from pediatric ICU, and 9 (15.8%) were from other inpatient services and outpatient clinics. A total of 17 (29.3%) of 58 clinical isolates were cultured from tracheal aspirate specimens, 10 (17.2%) of wound, 9 (15.5%) of urine, 5 (8.6%) of sputum, and 17 (29.3%) of other clinical materials.
4.1. Antibiotic Susceptibilities of Isolates
All of the 58 isolates were found to be resistant to imipenem and 43 of them were found to be resistant to meropenem with be disk diffusion test. Of the 58 isolates, 57 were found to be resistant and one of them was intermediate susceptible to imipenem; 43 of 58 isolates were found to be resistant and five were intermediate susceptible to meropenem with the VITEK 2 automated system. Resistance rates to other antipseudomonal antibiotics of 58 P. aeruginosa isolates resistant to carbapenems were found as amikasin 22%, gentamicin 29%, cefepime 32%, ceftazidime 39%, piperacilline 56%, ciprofloxacin 31% and levofloxacin 55% with VITEK 2. Of the 33 of our isolates were determined as MDR (multidrug resistant) and 25 (extremely drug resistant) were as XDR.
4.2. Determination of the MIC Values by the Gradient Test (the E-Test)
The E-test was performed to determine the MIC values of the isolates for imipenem and meropenem. It was found that the MIC values for imipenem were higher than the MIC values for meropenem. According to these MIC values, 34 isolates were found as resistant, 17 isolates were intermediate susceptible, and 7 isolates were susceptible to meropenem. The 57 isolates were found as resistant and only one isolate was intermediate susceptible to imipenem.
4.3. Investigation of the Presence of MBL by Phenotypic Tests
The CDT and the MBL E-test were applied to the isolates in this study. It was found that 16 (27%) isolates showed a positive result with the MBL E-test, and 37 (63%) isolates showed a positive result with the CDT (Figures 1 and 2, Table 2).
| CDT | MBL E-Test | |||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Pseudomonasaeruginosa | 37 (63) | 21 (27) | 16 (27) | 42 (73) |
aValues are expressed as No. (%).
4.4. MBL Molecular Analysis Results
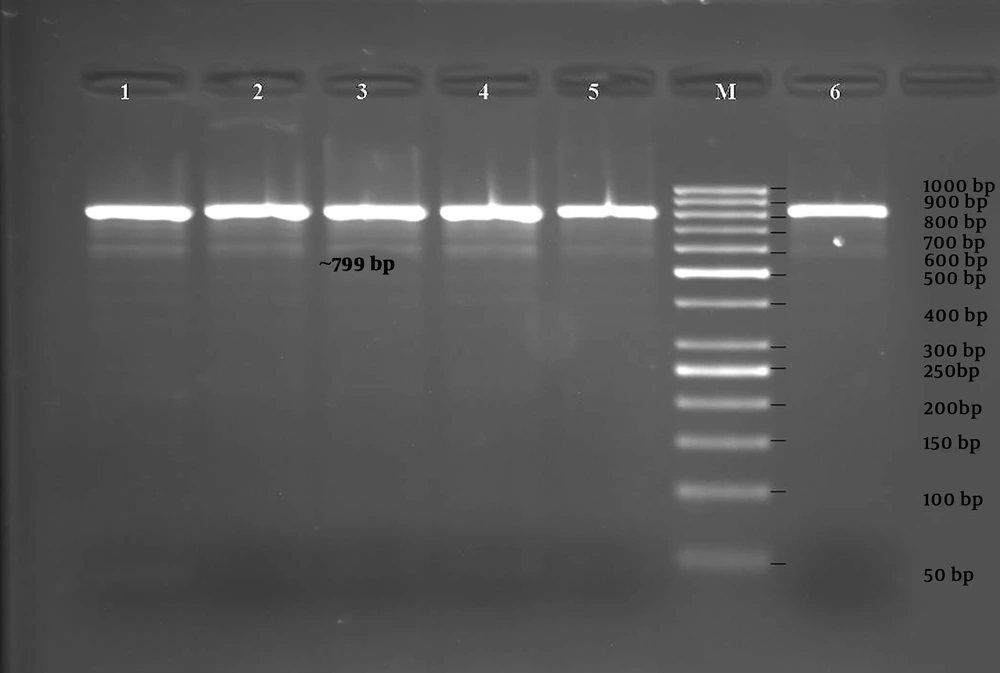
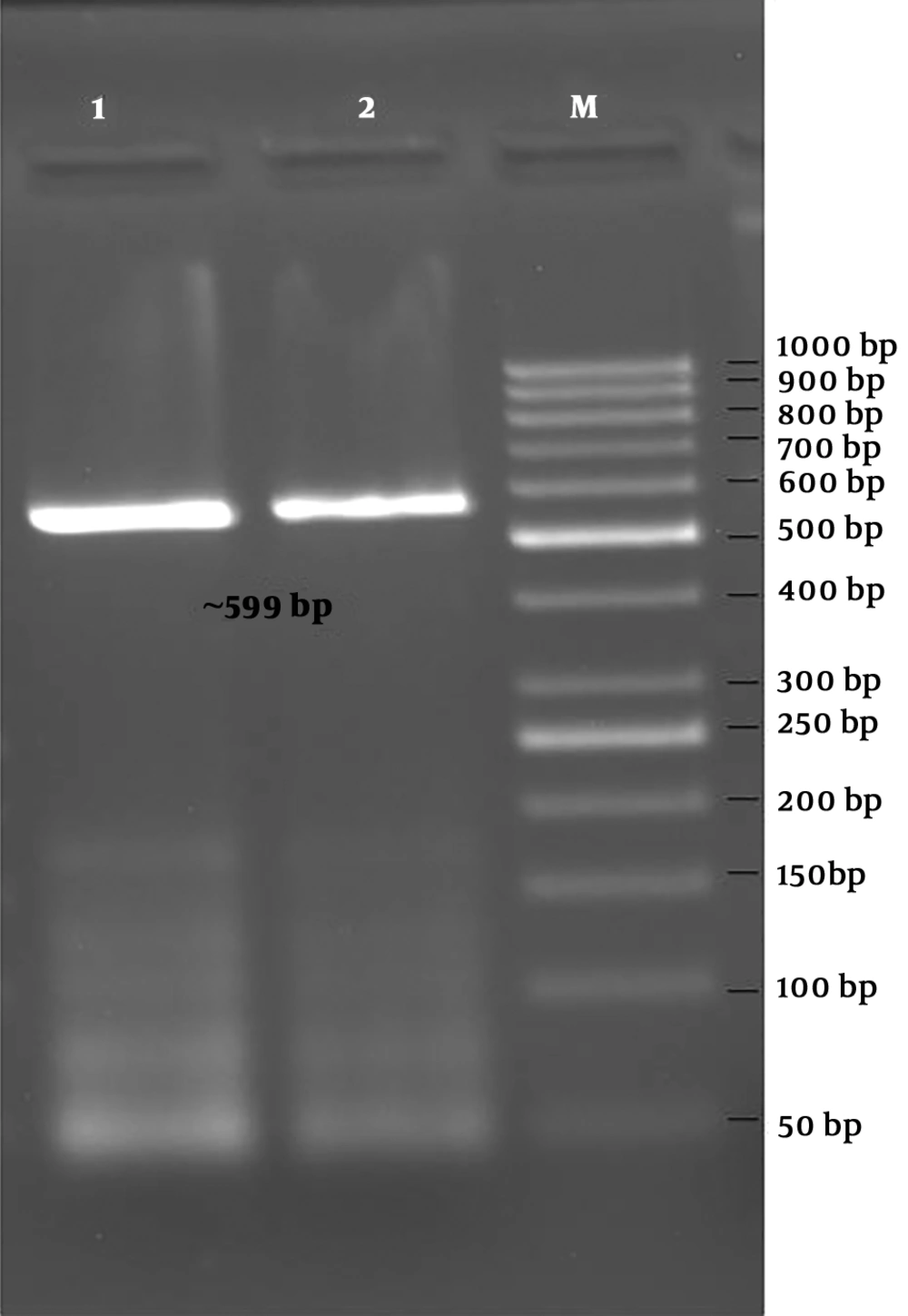
The PCR was performed using primer pairs specific for the blaIMP-1, blaIMP-2, blaVIM-1, blaVIM-2, blaGIM-1, and blaSPM genes. According to the results obtained by PCR, 6 isolates were positive for the blaVIM-1 gene (799 bp, Figure 3), and 2 isolates were positive for the blaGIM-1 gene (599 bp, Figure 4) (Table 3). blaIMP-1, blaIMP-2, blaVIM-2, blaSPM-type MBL gene regions were not detected by PZR in any of the isolates.
| PCR | ||
|---|---|---|
| MBL phenotypic tests | Positive (N) | Negative (N) |
| Positive | 8 | 29 |
| CDT | ||
| Negative | 0 | 20 |
| Positive | 7 | 9 |
| MBL E-test | ||
| Negative | 1 | 41 |
4.5. Evaluation of Sequence Analysis Data
Sequence analysis data for samples that were positive for the blaVIM-1 and blaGIM-1 genes were obtained chromatographically. These data were checked and corrected according to the peak lengths of the chromatogram waves in the Chromas DNA sequence analysis program. Moreover, the last consensus was recorded in series after the unreadable sequences in the end parts were clipped. Sequence analysis data for the blaVIM-1 and blaGIM-1 genes were uploaded to the PubMed-BLAST program (the Basic Local Alignment Search Tool). They were compared and verified with the NCBI Reference Sequence published in the GenBank database. The bit-score, maximum identity (the number of identical sequences in comparison), and E values (statistical significance of comparison) were considered in the evaluation. The determination of the blaVIM-1 and blaGIM-1 genes were based on sequence matches, which gave similar and meaningful results over 96%. Verification were performed with over 98% and 96% base matching using a nucleotide sequence (Bank No: LC169578.1 and Bank No: CP008857.1) for the blaVIM-1 gene and for the blaGIM-1 gene respectively. GenBank access number taken for blaGIM-1 is MG954060.
5. Discussion
Pseudomonas aeruginosa is a microorganism commonly found in environments and it is a potential pathogen for community-acquired infections. Severe infections caused by P. aeruginosa are often acquired in the hospital and are the second most common cause of nosocomial pneumonia (2). It is one of the most important nosocomial infectious agent with high morbidity and mortality rates (20% - 50%) among Gram-negative pathogens (16). Pseudomonas aeruginosa has a natural resistance to most classes of antibiotics and can increase resistance to most of antibacterial agents even during treatment (2). According to the WHO data for Turkey in 2013, the 45% of hospital-acquired P. aeruginosa strains were isolated from ICU, and the carbapenem resistance rate was 33% (17). In our study, 41.3% of the isolates were isolated from the intensive care units and 37.9% of them were isolated from the respiratory tract samples (tracheal aspirate and sputum). Epidemiological studies have shown that infections with resistant P. aeruginosa increase morbidity-mortality rate, hospitalization time, chronic care and overall treatment cost of infection (2).
Carbapenems are the most effective beta-lactams in the treatment of P. aeruginosa infections because they exhibit high affinity for penicillin-binding protein, are stable against broad-spectrum betalactamases, and easily pass through the outer membrane (8). Carbapenem resistance in P. aeruginosa is mediated by the release of MBL, the loss of the OprD porin, and the excretion of the drug from the bacteria via efflux pumps. The release of carbapenemase from these is important because of the genes encoding to these enzymes are transferred via plasmids, transposons and spread among isolates (3, 18).
Early and accurate detection of carbapenemase-producing P. aeruginosa is necessary to prevent the propagation of carbapenemases across all Gram-negative organisms in healthcare settings, and accurate and cost-effective phenotypic assays are available to detect carbapenemase-producing P. aeruginosa (19). In our study, the Gradient Test was performed to confirm carbapenem resistance. The AST results obtained for imipenem by the Gradient Test were found to be the same with the disc diffusion method and automated system. However, of the 24 isolates resistant to meropenem by the other two methods, 17 were intermediate susceptible to meropenem, and 7 were susceptible to meropenem. In our country, Ogunc et al. (20) investigated the imipenem and meropenem susceptibilities of P. aeruginosa and Acinetobacter baumannii strains, which were found to be resistant or less susceptible to imipenem and/or meropenem by the BD Phoenix system, with the disc diffusion method, the E-test, and the broth microdilution method. They found that all 51 P. aeruginosa isolates were resistant or less susceptible to imipenem and/or meropenem by the broth microdilution method. When the broth microdilution method was taken as a reference, the E-test had a 98% compatibility level.
In a review analyzing the epidemiology of MBL-producing P. aeruginosa isolates, the data collected from 50 countries including Turkey were examined. The countries with the lowest carbapenem resistance rate among clinical isolates of P. aeruginosa were Canada and the Dominican Republic (3% and 8%, respectively). The countries with the highest carbapenem resistance rate among clinical isolates of P. aeruginosa were Brazil, Peru, Costa Rica, Russia, Greece, Poland, Iran, and Saudi Arabia (ranging between 50% - 75.3%) (8). According to the WHO’s data for Turkey in 2013, the carbapenem resistance rate of hospital-acquired P. aeruginosa strains in our country was reported to be 33% (17). The strains of 720 P. aeruginosa were isolated from 392 patients in our hospital in 2015. When the recurrent isolates were excluded and the first isolate obtained from each patient was evaluated, 24.7% and 26.4% of them were resistant to imipenem and to meropenem respectively.
The PCR is the Gold standard method in demonstrating the presence of MBL. However, phenotypic tests are also needed because PCR is expensive and requires experienced staff. The CDT, the double disc synergy test, the Modified Hodge test, and the MBL Gradient Test are used for this purpose (9, 21). The imipenem-EDTA CDT is one of the phenotypic tests based on the observation of the synergistic effect of metal chelators with beta-lactam antibiotics used in MBL detection. In a study of Moosavian et al., they showed that 110 (90%) of the 122 imipenem resistant P. aeruginosa isolates, 67 of which had the MBL gene by the PCR, were found to be positive by the CDT (4). In our country, in a study of Ozgumus et al. performed with the 33 P. aeruginosa isolates confirmed to be resistant to at least one carbapenem. MBL production was detected in 29 (88%) of the isolates by the MBL strip which contained IPM-EDTA (22). In a study conducted in our hospital in 2011, the presence of MBL was investigated by phenotypic and genotypic methods in 29 carbapenem- resistant P. aeruginosa isolates, which were thought to be the cause of nosocomial infection. A total of 6 (20.7%) isolates were found to be positive by the MBL E-test (publicly available data) (23).
A study was conducted to investigate the incidence and global distribution of MBL in P. aeruginosa and Enterobacteriaceae isolates with the participation of 40 countries, including Turkey, between 2012 and 2014. It was found that 308 (3.8%) of the P. aeruginosa isolates in the study had MBL genes. Of the genes found to be positive in P. aeruginosa, 87.7% were blaVIM, 11% were blaIMP, and 1% was blaNDM. In addition, blaVIM-2 was the most frequently detected VIM-type (24). The blaVIM-5, which was detected in P. aeruginosa isolate in a study of Bahar et al. in 2004, is the first detected MBL gene in our country (25). Yakupogullari et al. described a P. aeruginosa isolate expressing blaVIM-2 and PER-1 in our country in 2007 (26). In a study of Ozgumus et al. conducted in 100 P. aeruginosa isolates in 2007, MBL was detected in a total of 10 isolates, including blaVIM in 1 (1%) and blaIMP in 9 (9%) (27).
Yilmaz et al. found that of the 38 carbapenem resistant isolates, 7 (18%) had blaVIM-2 and 1 (2%) had blaIMP-9 (28). In a study of Er et al. made in 195 P. aeruginosa isolates including 95 carbapenem-resistant isolates, it was found that 4 (2%) were positive for blaVIM-2, 2 (1%) were positive for blaIMP-1, and 26 (13%) were positive for blaGES-1 (29). In a study of Malkocoglu et al. performed in 84 carbapenem-resistant P. aeruginosa isolates, it was found that 1 (1.1%) was positive for blaVIM-1, 1 (1.1%) was positive for blaVIM-2, and 1 (1.1%) was positive for blaGES-5 (15). In a master thesis study conducted in our country in 2011, the presence of the blaIMP-1, blaIMP-2, blaVIM-1, and blaVIM-2 genes were investigated in 29 carbapenem-resistant P. aeruginosa isolates, which were thought to be the cause of nosocomial infection. A total of 11 (37.9%) were found to be positive for the blaVIM-1 gene (23).
In international studies conducted in recent years, the positivity rates of MBL genes vary between 0.9% and 68% (3, 12, 30). In studies performed in our country, while MBL genes were never detected in some regions, it was found to be as high as 37.9% in some other regions (18, 22, 23, 28, 29). Pseudomonas aeruginosa has a large number of gene regions responsible for MBL resistance and new ones are added to this list. The specificity of the primers used in molecular studies and the emergence of new variants affect the results and different results can be obtained. Therefore, in the isolates with MBL activity by phenotypic tests, resistance gene regions could not be detected at the molecular level. In our study, 63% MBL production was detected with CDT and 27% with MBL E-test. The presence of MBL genes was determined in 13.7% of isolates by PCR and the most common MBL variant was found as blaVIM-1 gene.
In our study, 8 strains were positive for an MBL gene by PCR and all of them were found as positive by phenotypic methods. However, the one isolate which has positive MBL activity by PCR was found to be positive MBL by CDT and negative by MBL-E test. Targeted gene region by PCR was not detected in 29 isolates of CDT positive and 9 isolates of MBL-E test positive. The development of carbapenem resistance among P. aeruginosa strains is multifactorial. For example, plasmid or integron-mediated carbapenemases, increased expression of efflux systems, reduced porin expression and increased chromosomal cephalosporinase activity have all been defined as contributory factors (31). The reason of the discrepancy in our study has been thought to might be other metallo-beta-lactamase genes in these isolates.
In our study, blaGIM-1 was found to be positive in 2 isolates, unlike the other studies. GIM (German imipenemase) was first described in a P. aeruginosa isolate in Germany in 2004 (32). It has been demonstrated that this gene, which has also been detected in Enterobacter cloacae, Serratia marcescens, and Acinetobacter pittii in the following years, is carried by plasmids but is not transferable (33-36). In studies conducted in Turkey, the GIM gene has not been detected in P. aeruginosa. In the light of current literature data, this study is the first study showing the blaGIM-1 gene in P. aeruginosa in Turkey. For this reason, it is important to verify the gene regions that are positive in molecular studies by sequence analysis. Moreover, we suggest molecular surveillance of MBL-producing isolates, e.g., RAPD, PFGE to increase the spread resistant clones in healthcare centers.
5.1. Conclusions
In brief, P. aeruginosa has developed resistance to many anti-pseudomonal antibiotics, including carbapenems. It is important to follow up the AST results and the resistance mechanisms in hospitals because carbapenem resistance in P. aeruginosa restricts treatment options, and resistance genes can be transferred between strains. Determining the presence of MBL in the laboratory by fast and reliable methods will ensure that appropriate infection control measures are taken to prevent spread of these strains in hospitals. For this purpose, phenotypic tests that are cost effective for detecting MBL in P. aeruginosa can be used as screening tests in laboratories where molecular tests cannot be applied. Furthermore, the investigation of MBL resistance genes would provide the collection of national epidemiological data.